Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy
- PMID: 10516049
- PMCID: PMC112975
- DOI: 10.1128/JVI.73.11.9404-9412.1999
Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy
Abstract
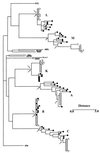
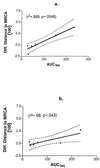
In human immunodeficiency virus (HIV)-infected patients treated with potent antiretroviral therapy, the persistence of latently infected cells may reflect the long decay half-life of this cellular reservoir or ongoing viral replication at low levels with continuous replenishment of the population or both. To address these possibilities, sequences encompassing the C2 and V3 domains of HIV-1 env were analyzed from virus present in baseline plasma and from viral isolates obtained after 2 years of suppressive therapy in six patients. The presence of sequence changes consistent with evolution was demonstrated for three subjects and correlated with less complete suppression of viral replication, as indicated by the rapidity of the initial virus load decline or the intermittent reappearance of even low levels of detectable viremia. Together, these results provide evidence for ongoing replication. In the remaining three patients, virus recovered after 2 years of therapy was either genotypically contemporary with or ancestral to virus present in plasma 2 years before, indicating that virus recovery had indeed resulted from activation of latently infected cells.
Figures


References
-
- Cavert W, Notermans D W, Staskus K, Wietgrefe S, Zupancic M, Gebhard K, Henry K, Zhang Z Q, Mills R, McDade H, Goudsmit J, Danner S A, Haase A T. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–964. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

