Requirements for RNA replication of a poliovirus replicon by coxsackievirus B3 RNA polymerase
- PMID: 10516050
- PMCID: PMC112976
- DOI: 10.1128/JVI.73.11.9413-9421.1999
Requirements for RNA replication of a poliovirus replicon by coxsackievirus B3 RNA polymerase
Abstract
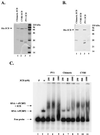
A chimeric poliovirus type 1 (PV1) genome was constructed in which the 3D RNA polymerase (3D(pol)) coding sequences were replaced with those from coxsackievirus B3 (CVB3). No infectious virus was produced from HeLa cells transfected with the chimeric RNA. Processing of the PV1 capsid protein precursor was incomplete, presumably due to inefficient recognition of the P1 protein substrate by the chimeric 3CD proteinase containing CVB3 3D sequences. The ability of the chimeric RNA to replicate in the absence of capsid formation was measured after replacement of the P1 region with a luciferase reporter gene. No RNA synthesis was detected, despite efficient production of enzymatically active 3D(pol) from the 3D portion of the chimeric 3CD. The chimeric 3CD protein was unable to efficiently bind to the cloverleaf-like structure (CL) at the 5' end of PV1 RNA, which has been demonstrated previously to be required for viral RNA synthesis. The CVB3 3CD protein bound the PV1 CL as well as PV1 3CD. An additional chimeric PV1 RNA that contained CVB3 3CD sequences also failed to produce virus after transfection. Since processing of PV1 capsid protein precursors by the CVB3 3CD was again incomplete, a luciferase-containing replicon was also analyzed for RNA replication. The 3CD chimera replicated at 33 degrees C, but not at 37 degrees C. Replacement of the PV1 5'-terminal CL with that of CVB3 did not rescue the temperature-sensitive phenotype. Thus, there is an essential interaction(s) between 3CD and other viral P2 or P3 protein products required for efficient RNA replication which is not fully achieved between proteins from the two different members of the same virus genus.
Figures






References
-
- Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. - PubMed
-
- Bell, Y. C., J. H. C. Nguyen, and B. L. Semler. Unpublished observations.
-
- Bienz K, Egger D, Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987;160:220–226. - PubMed
-
- Blair W S, Nguyen J H, Parsley T B, Semler B L. Mutations in the poliovirus 3CD proteinase S1-specificity pocket affect substrate recognition and RNA binding. Virology. 1996;218:1–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

