Plant succinic semialdehyde dehydrogenase. Cloning, purification, localization in mitochondria, and regulation by adenine nucleotides
- PMID: 10517851
- PMCID: PMC59422
- DOI: 10.1104/pp.121.2.589
Plant succinic semialdehyde dehydrogenase. Cloning, purification, localization in mitochondria, and regulation by adenine nucleotides
Abstract
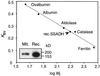
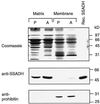
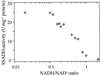
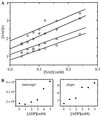
Succinic semialdehyde dehydrogenase (SSADH) is one of three enzymes constituting the gamma-aminobutyric acid shunt. We have cloned the cDNA for SSADH from Arabidopsis, which we designated SSADH1. SSADH1 cDNA encodes a protein of 528 amino acids (56 kD) with high similarity to SSADH from Escherichia coli and human (>59% identity). A sequence similar to a mitochondrial protease cleavage site is present 33 amino acids from the N terminus, indicating that the mature mitochondrial protein may contain 495 amino acids (53 kD). The native recombinant enzyme and the plant mitochondrial protein have a tetrameric molecular mass of 197 kD. Fractionation of plant mitochondria revealed its localization in the matrix. The purified recombinant enzyme showed maximal activity at pH 9.0 to 9.5, was specific for succinic semialdehyde (K(0.5) = 15 microM), and exclusively used NAD+ as a cofactor (Km = 130 +/- 77 microM). NADH was a competitive inhibitor with respect to NAD+ (Ki = 122 +/- 86 microM). AMP, ADP, and ATP inhibited the activity of SSADH (Ki = 2.5-8 mM). The mechanism of inhibition was competitive for AMP, noncompetitive for ATP, and mixed competitive for ADP with respect to NAD+. Plant SSADH may be responsive to mitochondrial energy charge and reducing potential in controlling metabolism of gamma-aminobutyric acid.
Figures



Similar articles
-
Plant succinic semialdehyde dehydrogenase: dissection of nucleotide binding by surface plasmon resonance and fluorescence spectroscopy.Biochemistry. 2000 Aug 22;39(33):10110-7. doi: 10.1021/bi000589e. Biochemistry. 2000. PMID: 10955999
-
Molecular cloning of the mature NAD(+)-dependent succinic semialdehyde dehydrogenase from rat and human. cDNA isolation, evolutionary homology, and tissue expression.J Biol Chem. 1995 Jan 6;270(1):461-7. doi: 10.1074/jbc.270.1.461. J Biol Chem. 1995. PMID: 7814412
-
Characterization of the recombinant succinic semi-aldehyde dehydrogenase from Saccharomyces cerevisiae.Yeast. 2014 Oct;31(10):411-20. doi: 10.1002/yea.3035. Epub 2014 Sep 8. Yeast. 2014. PMID: 25092794
-
Vigabatrin and newer interventions in succinic semialdehyde dehydrogenase deficiency.Ann Neurol. 2003;54 Suppl 6:S66-72. doi: 10.1002/ana.10626. Ann Neurol. 2003. PMID: 12891656 Review.
-
[Role of adenine mono- and dinucleotides in ammonia formation in brain tissue].Vopr Biokhim Mozga. 1975;10:5-32. Vopr Biokhim Mozga. 1975. PMID: 186942 Review. Russian.
Cited by
-
Inhibition of aconitase in citrus fruit callus results in a metabolic shift towards amino acid biosynthesis.Planta. 2011 Sep;234(3):501-13. doi: 10.1007/s00425-011-1411-2. Epub 2011 Apr 28. Planta. 2011. PMID: 21528417
-
Structural basis for a cofactor-dependent oxidation protection and catalysis of cyanobacterial succinic semialdehyde dehydrogenase.J Biol Chem. 2013 May 31;288(22):15760-70. doi: 10.1074/jbc.M113.460428. Epub 2013 Apr 15. J Biol Chem. 2013. PMID: 23589281 Free PMC article.
-
Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants.Proc Natl Acad Sci U S A. 2003 May 27;100(11):6843-8. doi: 10.1073/pnas.1037532100. Epub 2003 May 9. Proc Natl Acad Sci U S A. 2003. PMID: 12740438 Free PMC article.
-
Analysis of the Arabidopsis mitochondrial proteome.Plant Physiol. 2001 Dec;127(4):1711-27. Plant Physiol. 2001. PMID: 11743115 Free PMC article.
-
Comparative study of the aldehyde dehydrogenase (ALDH) gene superfamily in the glycophyte Arabidopsis thaliana and Eutrema halophytes.Ann Bot. 2015 Feb;115(3):465-79. doi: 10.1093/aob/mcu152. Epub 2014 Aug 1. Ann Bot. 2015. PMID: 25085467 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Baum G, Chen Y, Arazi T, Takutsuji H, Fromm H. A plant glutamate decarboxylase containing a calmodulin binding domain: cloning, sequence, and functional analysis. J Biol Chem. 1993;268:19610–19617. - PubMed
-
- Bisswanger H. Enzymkinetik. Weinheim, Germany: VHC; 1994.
-
- Blaner WS, Churchich J. The binding of NADH to succinic semialdehyde dehydrogenase. Eur J Biochem. 1980;109:431–437. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

