Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus)
- PMID: 10518550
- PMCID: PMC18386
- DOI: 10.1073/pnas.96.21.11910
Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus)
Abstract
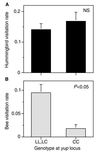
A paradigm of evolutionary biology is that adaptation and reproductive isolation are caused by a nearly infinite number of mutations of individually small effect. Here, we test this hypothesis by investigating the genetic basis of pollinator discrimination in two closely related species of monkeyflowers that differ in their major pollinators. This system provides a unique opportunity to investigate the genetic architecture of adaptation and speciation because floral traits that confer pollinator specificity also contribute to premating reproductive isolation. We asked: (i) What floral traits cause pollinator discrimination among plant species? and (ii) What is the genetic basis of these traits? We examined these questions by using data obtained from a large-scale field experiment where genetic markers were employed to determine the genetic basis of pollinator visitation. Observations of F2 hybrids produced by crossing bee-pollinated Mimulus lewisii with hummingbird-pollinated Mimulus cardinalis revealed that bees preferred large flowers low in anthocyanin and carotenoid pigments, whereas hummingbirds favored nectar-rich flowers high in anthocyanins. An allele that increases petal carotenoid concentration reduced bee visitation by 80%, whereas an allele that increases nectar production doubled hummingbird visitation. These results suggest that genes of large effect on pollinator preference have contributed to floral evolution and premating reproductive isolation in these monkeyflowers. This work contributes to growing evidence that adaptation and reproductive isolation may often involve major genes.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

