The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms
- PMID: 10521401
- PMCID: PMC317077
- DOI: 10.1101/gad.13.19.2570
The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms
Abstract
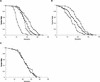
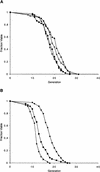
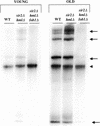
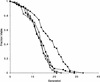
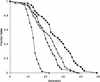
The SIR genes are determinants of life span in yeast mother cells. Here we show that life span regulation by the Sir proteins is independent of their role in nonhomologous end joining. The short life span of a sir3 or sir4 mutant is due to the simultaneous expression of a and alpha mating-type information, which indirectly causes an increase in rDNA recombination and likely increases the production of extrachromosomal rDNA circles. The short life span of a sir2 mutant also reveals a direct failure to repress recombination generated by the Fob1p-mediated replication block in the rDNA. Sir2p is a limiting component in promoting yeast longevity, and increasing the gene dosage extends the life span in wild-type cells. A possible role of the conserved SIR2 in mammalian aging is discussed.
Figures

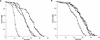

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
