Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia
- PMID: 10523559
- PMCID: PMC85701
- DOI: 10.1128/JCM.37.11.3594-3600.1999
Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia
Abstract
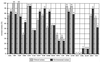
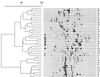
While the gram-negative bacterium Stenotrophomonas maltophilia is used in biotechnology (e.g., for biological control of plant pathogens and for bioremediation), the number of S. maltophilia diseases in humans has dramatically increased in recent years. A total of 40 S. maltophilia isolates from clinical and environmental sources (plant associated and water) was investigated to determine the intraspecies diversity of the group and to determine whether or not the strains could be grouped based on the source of isolation. The isolates were investigated by phenotypic profiling (enzymatic and metabolic activity and antibiotic resistance patterns) and by molecular methods such as temperature-gradient gel electrophoresis of the 16S rRNA gene fragment, PCR fingerprinting with BOX primers, and pulsed-field gel electrophoresis (PFGE) after digestion with DraI. Results of the various methods revealed high intraspecies diversity. PFGE was the most discriminatory method for typing S. maltophilia when compared to the other molecular methods. The environmental strains of S. maltophilia were highly resistant to antibiotics, and the resistance profile pattern of the strains was not dependent on their source of isolation. Computer-assisted cluster analysis of the phenotypic and genotypic features did not reveal any clustering patterns for either clinical or environmental isolates.
Figures


References
-
- Arpi M, Victor M A, Mortensen I, Gottschau A, Bruun B. In vitro susceptibility of 124 Xanthomonas maltophilia (Stenotrophomonas maltophilia) isolates: comparison of the agar dilution method with the E-test and two agar diffusion methods. APMIS. 1996;104:108–114. - PubMed
-
- Berg G, Ballin G. Bacterial antagonists of Verticillium dahliae KLEB. J Phytopathol. 1994;141:99–110.
-
- Berg G, Knaape C, Ballin G, Seidel D. Biological control of Verticillium dahliae KLEB by naturally occurring rhizosphere bacteria. Arch Phytopathol Dis Prot. 1994;29:249–262.
-
- Berg G, Marten P, Ballin G. Stenotrophomonas maltophilia in the rhizosphere of oilseed rape—occurrence, characterization and interaction with phytopathogenic fungi. Microbiol Res. 1996;151:19–27.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

