A sampling of the yeast proteome
- PMID: 10523624
- PMCID: PMC84729
- DOI: 10.1128/MCB.19.11.7357
A sampling of the yeast proteome
Abstract
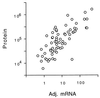
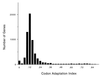
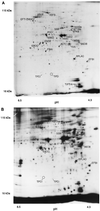
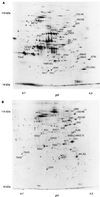
In this study, we examined yeast proteins by two-dimensional (2D) gel electrophoresis and gathered quantitative information from about 1,400 spots. We found that there is an enormous range of protein abundance and, for identified spots, a good correlation between protein abundance, mRNA abundance, and codon bias. For each molecule of well-translated mRNA, there were about 4,000 molecules of protein. The relative abundance of proteins was measured in glucose and ethanol media. Protein turnover was examined and found to be insignificant for abundant proteins. Some phosphoproteins were identified. The behavior of proteins in differential centrifugation experiments was examined. Such experiments with 2D gels can give a global view of the yeast proteome.
Figures


References
-
- Boucherie H, Sagliocco F, Joubert R, Maillet I, Labarre J, Perrot M. Two-dimensional gel protein database of Saccharomyces cerevisiae. Electrophoresis. 1996;17:1683–1699. - PubMed
-
- Elliott B, Futcher B. Stress resistance of yeast cells is largely independent of cell cycle phase. Yeast. 1993;9:33–42. - PubMed
-
- Entian K D, Meurer B, Kohler H, Mann K H, Mecke D. Studies on the regulation of enolases and compartmentation of cytosolic enzymes in Saccharomyces cerevisiae. Biochim Biophys Acta. 1987;923:214–221. - PubMed
-
- Ganzhorn A J, Green D W, Hershey A D, Gould R M, Plapp B V. Kinetic characterization of yeast alcohol dehydrogenases. Amino acid residue 294 and substrate specificity. J Biol Chem. 1987;262:3754–3761. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
