PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins
- PMID: 10523646
- PMCID: PMC84774
- DOI: 10.1128/MCB.19.11.7577
PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins
Abstract
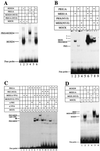
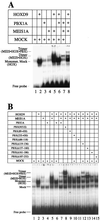
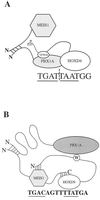
HOX, PBX, and MEIS transcription factors bind DNA through a homeodomain. PBX proteins bind DNA cooperatively as heterodimers with MEIS family members and also with HOX proteins from paralog groups 1 to 10. MEIS proteins cooperatively bind DNA with ABD-B class HOX proteins of groups 9 and 10. Here, we examine aspects of dimeric and higher-order interactions between these three homeodomain classes. The most significant results can be summarized as follows. (i) Most of PBX N terminal to the homeodomain is required for efficient cooperative binding with HOXD4 and HOXD9. (ii) MEIS and PBX proteins form higher-order complexes on a heterodimeric binding site. (iii) Although MEIS does not cooperatively bind DNA with ANTP class HOX proteins, it does form a trimer as a non-DNA-binding partner with DNA-bound PBX-HOXD4. (iv) The N terminus of HOXD4 negatively regulates trimer formation. (v) MEIS forms a similar trimer with DNA-bound PBX-HOXD9. (vi) A related trimer (where MEIS is a non-DNA-binding partner) is formed on a transcriptional promoter within the cell. (vii) We observe an additional trimer class involving non-DNA-bound PBX and DNA-bound MEIS-HOXD9 or MEIS-HOXD10 heterodimers that is enhanced by mutation of the PBX homeodomain. (viii) In this latter trimer, PBX is likely to contact both MEIS and HOXD9/D10. (ix) The stability of DNA binding by all trimers is enhanced relative to the heterodimers. These findings suggest novel functions for PBX and MEIS in modulating the function of DNA-bound MEIS-HOX and PBX-HOX heterodimers, respectively.
Figures






References
-
- Andrew S M, Titus J A. Fragmentation of immunoglobulin G. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1992.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
