The levels of the bancal product, a Drosophila homologue of vertebrate hnRNP K protein, affect cell proliferation and apoptosis in imaginal disc cells
- PMID: 10523673
- PMCID: PMC84859
- DOI: 10.1128/MCB.19.11.7846
The levels of the bancal product, a Drosophila homologue of vertebrate hnRNP K protein, affect cell proliferation and apoptosis in imaginal disc cells
Abstract
We have characterized the Drosophila bancal gene, which encodes a Drosophila homologue of the vertebrate hnRNP K protein. The bancal gene is essential for the correct size of adult appendages. Reduction of appendage size in bancal mutant flies appears to be due mainly to a reduction in the number of cell divisions in the imaginal discs. Transgenes expressing Drosophila or human hnRNP K are able to rescue weak bancal phenotype, showing the functional similarity of these proteins in vivo. High levels of either human or Drosophila hnRNP K protein in imaginal discs induces programmed cell death. Expression of the antiapoptotic P35 protein suppresses this phenotype in the eye, suggesting that apoptosis is the major cellular defect caused by overexpression of K protein. Finally, the human K protein acts as a negative regulator of bancal gene expression. We propose that negative autoregulation limits the level of Bancal protein produced in vivo.
Figures
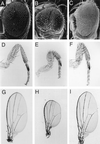




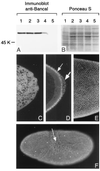
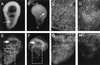
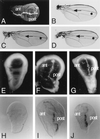


References
-
- Alexandre E, Graba Y, Fasano L, Gallet A, Perrin L, De Zulueta P, Pradel J, Kerridge S, Jacq B. The Drosophila Teashirt homeotic protein is a DNA-binding protein and modulo, a HOM C regulated modifier of variegation, is a likely candidate for being a direct target gene. Mech Dev. 1996;59:191–204. - PubMed
-
- Amrein H, Gorman M, Nothiger R. The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell. 1988;55:1025–1035. - PubMed
-
- Baker N E, Rubin G M. Ellipse mutations in the Drosophila homologue of the EGF receptor affect pattern formation, cell division, and cell death in eye imaginal discs. Dev Biol. 1992;150:381–396. - PubMed
-
- Bell L R, Maine E M, Schedl P, Cline T W. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell. 1988;55:1037–1046. - PubMed
-
- Bellen H J, Kooyer S, D’Evelyn D, Pearlman J. The Drosophila couch potato protein is expressed in nuclei of peripheral neuronal precursors and shows homology to RNA-binding proteins. Genes Dev. 1992;6:2125–2136. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
