Novel roles for saccharomyces cerevisiae mitotic spindle motors
- PMID: 10525539
- PMCID: PMC2174232
- DOI: 10.1083/jcb.147.2.335
Novel roles for saccharomyces cerevisiae mitotic spindle motors
Abstract
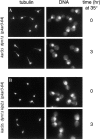
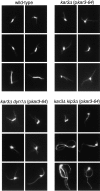
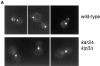
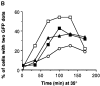
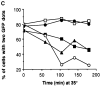
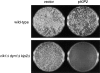
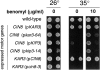
The single cytoplasmic dynein and five of the six kinesin-related proteins encoded by Saccharomyces cerevisiae participate in mitotic spindle function. Some of the motors operate within the nucleus to assemble and elongate the bipolar spindle. Others operate on the cytoplasmic microtubules to effect spindle and nuclear positioning within the cell. This study reveals that kinesin-related Kar3p and Kip3p are unique in that they perform roles both inside and outside the nucleus. Kar3p, like Kip3p, was found to be required for spindle positioning in the absence of dynein. The spindle positioning role of Kar3p is performed in concert with the Cik1p accessory factor, but not the homologous Vik1p. Kar3p and Kip3p were also found to overlap for a function essential for the structural integrity of the bipolar spindle. The cytoplasmic and nuclear roles of both these motors could be partially substituted for by the microtubule-destabilizing agent benomyl, suggesting that these motors perform an essential microtubule-destabilizing function. In addition, we found that yeast cell viability could be supported by as few as two microtubule-based motors: the BimC-type kinesin Cin8p, required for spindle structure, paired with either Kar3p or Kip3p, required for both spindle structure and positioning.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

