Expression and characterization of the Mycobacterium tuberculosis serine/threonine protein kinase PknB
- PMID: 10531215
- PMCID: PMC96941
- DOI: 10.1128/IAI.67.11.5676-5682.1999
Expression and characterization of the Mycobacterium tuberculosis serine/threonine protein kinase PknB
Abstract
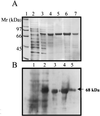
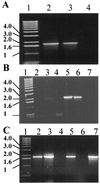
PknB is a member of the newly discovered eukaryotic-like protein serine/threonine kinase (PSTK) family of proteins. The pknB gene was cloned and expressed in Escherichia coli. The active recombinant protein was purified and shown to be reactive with antiphosphoserine antibodies, as well as with antibodies to the phosphorylated eukaryotic Ser/Thr kinases mitogen-activated protein kinase kinase 3 and 6, P38, and Creb. In vitro kinase assays demonstrated that PknB is a functional kinase that is autophosphorylated on serine/threonine residues and is also able to phosphorylate the peptide substrate myelin basic protein. Analysis of pknB expression in Mycobacterium tuberculosis indicates the presence of pknB mRNA in (i) organisms grown in vitro in bacteriological media, (ii) a murine macrophage in vitro infection model, and (iii) in vivo alveolar macrophages from a patient with tuberculosis.
Figures


References
-
- Asoh S, Matsuzawa H, Ishino F, Strominger J L, Matsuhashi M, Ohta T. Nucleotide sequence of the pbpA gene and characteristics of the deduced amino acid sequence of penicillin-binding protein 2 of Escherichia coli K12. Eur J Biochem. 1986;160:231–238. - PubMed
-
- Av-Gay Y, Davies J. Components of eukaryotic-like protein signaling pathways in Mycobacterium tuberculosis. Microb Comp Genomics. 1997;2:63–73.
-
- Brozna J P, Horan M, Rademacher J M, Pabst K M, Pabst M J. Monocyte responses to sulfatide from Mycobacterium tuberculosis: inhibition of priming for enhanced release of superoxide, associated with increased secretion of interleukin-1 and tumor necrosis factor alpha, and altered protein phosphorylation. Infect Immun. 1991;59:2542–2548. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

