Compromised disease resistance in saponin-deficient plants
- PMID: 10536024
- PMCID: PMC23166
- DOI: 10.1073/pnas.96.22.12923
Compromised disease resistance in saponin-deficient plants
Abstract
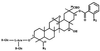
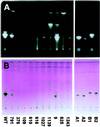
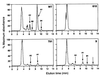
Saponins are glycosylated plant secondary metabolites found in many major food crops [Price, K. R., Johnson, I. T. & Fenwick, G. R. (1987) CRC Crit. Rev. Food Sci. Nutr. 26, 27-133]. Because many saponins have potent antifungal properties and are present in healthy plants in high concentrations, these molecules may act as preformed chemical barriers to fungal attack. The isolation of plant mutants defective in saponin biosynthesis represents a powerful strategy for evaluating the importance of these compounds in plant defense. The oat root saponin avenacin A-1 fluoresces under ultraviolet illumination [Crombie, L., Crombie, W. M. L. & Whiting, D. A. (1986) J. Chem. Soc. Perkins 1, 1917-1922], a property that is extremely rare among saponins. Here we have exploited this fluorescence to isolate saponin-deficient (sad) mutants of a diploid oat species, Avena strigosa. These sad mutants are compromised in their resistance to a variety of fungal pathogens, and a number of lines of evidence suggest that this compromised disease resistance is a direct consequence of saponin deficiency. Because saponins are widespread throughout the plant kingdom, this group of secondary metabolites may have general significance as antimicrobial phytoprotectants.
Figures


References
-
- Schönbeck F, Schlösser E. In: Physiological Plant Pathology. Heitefuss R, Williams P H, editors. Berlin: Springer; 1976. pp. 653–678.
-
- Müller K O, Börger H. Arb Biol Reichsasnstalt Landw Forstw Berlin. 1940;23:189–223.
-
- Paxton J D. Phytopathol Z. 1981;101:106–109.
LinkOut - more resources
Full Text Sources
Other Literature Sources

