Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea
- PMID: 10542158
- PMCID: PMC94121
- DOI: 10.1128/JB.181.21.6591-6599.1999
Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea
Abstract
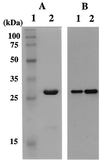
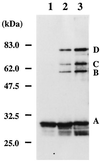
Proliferating cell nuclear antigen (PCNA) is an essential component of the DNA replication and repair machinery in the domain Eucarya. We cloned the gene encoding a PCNA homolog (PfuPCNA) from an euryarchaeote, Pyrococcus furiosus, expressed it in Escherichia coli, and characterized the biochemical properties of the gene product. The protein PfuPCNA stimulated the in vitro primer extension abilities of polymerase (Pol) I and Pol II, which are the two DNA polymerases identified in this organism to date. An immunological experiment showed that PfuPCNA interacts with both Pol I and Pol II. Pol I is a single polypeptide with a sequence similar to that of family B (alpha-like) DNA polymerases, while Pol II is a heterodimer. PfuPCNA interacted with DP2, the catalytic subunit of the heterodimeric complex. These results strongly support the idea that the PCNA homolog works as a sliding clamp of DNA polymerases in P. furiosus, and the basic mechanism for the processive DNA synthesis is conserved in the domains Bacteria, Eucarya, and Archaea. The stimulatory effect of PfuPCNA on the DNA synthesis was observed by using a circular DNA template without the clamp loader (replication factor C [RFC]) in both Pol I and Pol II reactions in contrast to the case of eukaryotic organisms, which are known to require the RFC to open the ring structure of PCNA prior to loading onto a circular DNA. Because RFC homologs have been found in the archaeal genomes, they may permit more efficient stimulation of DNA synthesis by archaeal DNA polymerases in the presence of PCNA. This is the first stage in elucidating the archaeal DNA replication mechanism.
Figures




References
-
- Arroyo M P, Downey K M, So A G, Wang T S-F. Schizosaccharomyces pombe proliferating cell nuclear antigen mutations affect DNA polymerase δ processivity. J Biol Chem. 1996;271:15971–15980. - PubMed
-
- Brown W C, Campbell J L. Interaction of proliferating cell nuclear antigen with yeast DNA polymerase δ. J Biol Chem. 1993;268:21706–21710. - PubMed
-
- Brush G S, Kelly T J. Mechanisms for replicating DNA. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 1–43.
-
- Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

