Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival
- PMID: 10542166
- PMCID: PMC94129
- DOI: 10.1128/JB.181.21.6656-6663.1999
Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival
Abstract
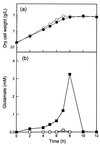
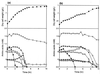
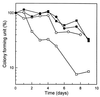
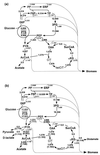
In order to study the physiological role of acetate metabolism in Escherichia coli, the growth characteristics of an E. coli W3100 pta mutant defective in phosphotransacetylase, the first enzyme of the acetate pathway, were investigated. The pta mutant grown on glucose minimal medium excreted unusual by-products such as pyruvate, D-lactate, and L-glutamate instead of acetate. In an analysis of the sequential consumption of amino acids by the pta mutant growing in tryptone broth (TB), a brief lag between the consumption of amino acids normally consumed was observed, but no such lag occurred for the wild-type strain. The pta mutant was found to grow slowly on glucose, TB, or pyruvate, but it grew normally on glycerol or succinate. The defective growth and starvation survival of the pta mutant were restored by the introduction of poly-beta-hydroxybutyrate (PHB) synthesis genes (phbCAB) from Alcaligenes eutrophus, indicating that the growth defect of the pta mutant was due to a perturbation of acetyl coenzyme A (CoA) flux. By the stoichiometric analysis of the metabolic fluxes of the central metabolism, it was found that the amount of pyruvate generated from glucose transport by the phosphoenolpyruvate-dependent phosphotransferase system (PTS) exceeded the required amount of precursor metabolites downstream of pyruvate for biomass synthesis. These results suggest that E. coli excretes acetate due to the pyruvate flux from PTS and that any method which alleviates the oversupply of acetyl CoA would restore normal growth to the pta mutant.
Figures
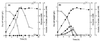
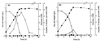
Similar articles
-
An insight into the role of phosphotransacetylase (pta) and the acetate/acetyl-CoA node in Escherichia coli.Microb Cell Fact. 2009 Oct 24;8:54. doi: 10.1186/1475-2859-8-54. Microb Cell Fact. 2009. PMID: 19852855 Free PMC article.
-
The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli.J Gen Microbiol. 1977 Oct;102(2):327-36. doi: 10.1099/00221287-102-2-327. J Gen Microbiol. 1977. PMID: 21941
-
Functional dissection of Escherichia coli phosphotransacetylase structural domains and analysis of key compounds involved in activity regulation.FEBS J. 2010 Apr;277(8):1957-66. doi: 10.1111/j.1742-4658.2010.07617.x. Epub 2010 Mar 8. FEBS J. 2010. PMID: 20236319
-
Control of carbon flux through enzymes of central and intermediary metabolism during growth of Escherichia coli on acetate.Curr Opin Microbiol. 2006 Apr;9(2):173-9. doi: 10.1016/j.mib.2006.02.002. Epub 2006 Mar 10. Curr Opin Microbiol. 2006. PMID: 16530464 Review.
-
Acetate Metabolism in Physiology, Cancer, and Beyond.Trends Cell Biol. 2019 Sep;29(9):695-703. doi: 10.1016/j.tcb.2019.05.005. Epub 2019 May 31. Trends Cell Biol. 2019. PMID: 31160120 Free PMC article. Review.
Cited by
-
Structural and functional studies suggest a catalytic mechanism for the phosphotransacetylase from Methanosarcina thermophila.J Bacteriol. 2006 Feb;188(3):1143-54. doi: 10.1128/JB.188.3.1143-1154.2006. J Bacteriol. 2006. PMID: 16428418 Free PMC article.
-
Repression of Escherichia coli PhoP-PhoQ signaling by acetate reveals a regulatory role for acetyl coenzyme A.J Bacteriol. 2003 Apr;185(8):2563-70. doi: 10.1128/JB.185.8.2563-2570.2003. J Bacteriol. 2003. PMID: 12670981 Free PMC article.
-
Inactivation of the Pta-AckA pathway impairs fitness of Bacillus anthracis during overflow metabolism.J Bacteriol. 2021 May 1;203(9):e00660-20. doi: 10.1128/JB.00660-20. Epub 2021 Feb 16. J Bacteriol. 2021. PMID: 33593944 Free PMC article.
-
L-Tryptophan Production in Escherichia coli Improved by Weakening the Pta-AckA Pathway.PLoS One. 2016 Jun 27;11(6):e0158200. doi: 10.1371/journal.pone.0158200. eCollection 2016. PLoS One. 2016. PMID: 27348810 Free PMC article.
-
Genetics and Physiology of Acetate Metabolism by the Pta-Ack Pathway of Streptococcus mutans.Appl Environ Microbiol. 2015 Aug;81(15):5015-25. doi: 10.1128/AEM.01160-15. Epub 2015 May 15. Appl Environ Microbiol. 2015. PMID: 25979891 Free PMC article.
References
-
- Amarsingham C R, Davis B D. Regulation of α-ketoglutarate dehydrogenase formation in Escherichia coli. J Biol Chem. 1965;240:3664–3668. - PubMed
-
- Atkinson M R, Ninfa A J. Role of GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol Microbiol. 1998;29:431–447. - PubMed
-
- Braunegg G, Sonnleitner B, Lafferty R M. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol. 1978;6:29–37.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

