The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons
- PMID: 10542180
- PMCID: PMC94143
- DOI: 10.1128/JB.181.21.6772-6778.1999
The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons
Abstract
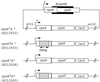
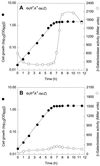
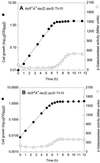
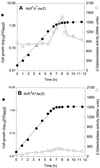
In Escherichia coli, the CpxRA two-component signal transduction system senses and responds to aggregated and misfolded proteins in the bacterial envelope. We show that CpxR-P (the phosphorylated form of the cognate response regulator) activates cpxRA expression in conjunction with RpoS, suggesting an involvement of the Cpx system in stationary-phase survival. Engagement of the CpxRA system in functions beyond protein management is indicated by several putative targets identified after a genomic screening for the CpxR-P recognition consensus sequence. Direct negative control of the newly identified targets motABcheAW (specifying motility and chemotaxis) and tsr (encoding the serine chemoreceptor) by CpxR-P was shown by electrophoretic mobility shift analysis and Northern hybridization. The results suggest that the CpxRA system plays a core role in an extensive stress response network in which the coordination of protein turnover and energy conservation may be the unifying element.
Figures



Similar articles
-
The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli.Genes Dev. 1997 May 1;11(9):1183-93. doi: 10.1101/gad.11.9.1183. Genes Dev. 1997. PMID: 9159399
-
Characterization of the CpxRA regulon in Haemophilus ducreyi.Infect Immun. 2010 Nov;78(11):4779-91. doi: 10.1128/IAI.00678-10. Epub 2010 Aug 30. Infect Immun. 2010. PMID: 20805330 Free PMC article.
-
The Cpx envelope stress response is controlled by amplification and feedback inhibition.J Bacteriol. 1999 Sep;181(17):5263-72. doi: 10.1128/JB.181.17.5263-5272.1999. J Bacteriol. 1999. PMID: 10464196 Free PMC article.
-
Transcriptional regulation of the bgl operon of Escherichia coli involves phosphotransferase system-mediated phosphorylation of a transcriptional antiterminator.J Cell Biochem. 1993 Jan;51(1):83-90. doi: 10.1002/jcb.240510115. J Cell Biochem. 1993. PMID: 7679391 Review. No abstract available.
-
Control of rpoS transcription in Escherichia coli and Pseudomonas: why so different?Mol Microbiol. 2003 Jul;49(1):1-9. doi: 10.1046/j.1365-2958.2003.03547.x. Mol Microbiol. 2003. PMID: 12823806 Review.
Cited by
-
Recombinant Human Peptidoglycan Recognition Proteins Reveal Antichlamydial Activity.Infect Immun. 2016 Jun 23;84(7):2124-2130. doi: 10.1128/IAI.01495-15. Print 2016 Jul. Infect Immun. 2016. PMID: 27160295 Free PMC article.
-
Maintaining Integrity Under Stress: Envelope Stress Response Regulation of Pathogenesis in Gram-Negative Bacteria.Front Cell Infect Microbiol. 2019 Sep 4;9:313. doi: 10.3389/fcimb.2019.00313. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31552196 Free PMC article. Review.
-
Negative regulation of DNA repair gene (ung) expression by the CpxR/CpxA two-component system in Escherichia coli K-12 and induction of mutations by increased expression of CpxR.J Bacteriol. 2004 Dec;186(24):8317-25. doi: 10.1128/JB.186.24.8317-8325.2004. J Bacteriol. 2004. PMID: 15576781 Free PMC article.
-
Phosphorylated CpxR restricts production of the RovA global regulator in Yersinia pseudotuberculosis.PLoS One. 2011;6(8):e23314. doi: 10.1371/journal.pone.0023314. Epub 2011 Aug 18. PLoS One. 2011. PMID: 21876746 Free PMC article.
-
The Three Streptomyces lividans HtrA-Like Proteases Involved in the Secretion Stress Response Act in a Cooperative Manner.PLoS One. 2016 Dec 15;11(12):e0168112. doi: 10.1371/journal.pone.0168112. eCollection 2016. PLoS One. 2016. PMID: 27977736 Free PMC article.
References
-
- Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and ςS in stationary phase. Mol Microbiol. 1994;13:265–272. - PubMed
-
- Cosma C L, Danese P N, Carlson J H, Silhavy T J, Snyder W B. Mutational activation by the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope-associated stresses. Mol Microbiol. 1995;18:491–505. - PubMed
-
- Danese P N, Silhavy T J. The ςE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997;11:1183–1193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

