Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis
- PMID: 10543800
- PMCID: PMC91658
- DOI: 10.1128/AEM.65.11.4887-4897.1999
Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis
Abstract
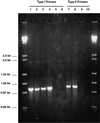
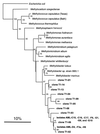
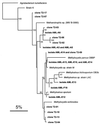
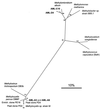
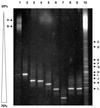
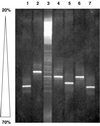
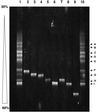
The diversity of the methanotrophic community in mildly acidic landfill cover soil was assessed by three methods: two culture-independent molecular approaches and a traditional culture-based approach. For the first of the molecular studies, two primer pairs specific for the 16S rRNA gene of validly published type I (including the former type X) and type II methanotrophs were identified and tested. These primers were used to amplify directly extracted soil DNA, and the products were used to construct type I and type II clone libraries. The second molecular approach, based on denaturing gradient gel electrophoresis (DGGE), provided profiles of the methanotrophic community members as distinguished by sequence differences in variable region 3 of the 16S ribosomal DNA. For the culturing studies, an extinction-dilution technique was employed to isolate slow-growing but numerically dominant strains. The key variables of the series of enrichment conditions were initial pH (4. 8 versus 6.8), air/CH(4)/CO(2) headspace ratio (50:45:5 versus 90:9:1), and concentration of the medium (1x nitrate minimal salts [NMS] versus 0.2x NMS). Screening of the isolates showed that the nutrient-rich 1x NMS selected for type I methanotrophs, while the nutrient-poor 0.2x NMS tended to enrich for type II methanotrophs. Partial sequencing of the 16S rRNA gene from selected clones and isolates revealed some of the same novel sequence types. Phylogenetic analysis of the type I clone library suggested the presence of a new phylotype related to the Methylobacter-Methylomicrobium group, and this was confirmed by isolating two members of this cluster. The type II clone library also suggested the existence of a novel group of related species distinct from the validly published Methylosinus and Methylocystis genera, and two members of this cluster were also successfully cultured. Partial sequencing of the pmoA gene, which codes for the 27-kDa polypeptide of the particulate methane monooxygenase, reaffirmed the phylogenetic placement of the four isolates. Finally, not all of the bands separated by DGGE could be accounted for by the clones and isolates. This polyphasic assessment of community structure demonstrates that much diversity among the obligate methane oxidizers has yet to be formally described.
Figures
References
-
- Amaral J A, Archambault C, Richards S R, Knowles R. Denitrification associated with group I and II methanotrophs in a gradient enrichment system. FEMS Microbiol Ecol. 1995;18:289–298.
-
- Bodrossy L, Holmes E M, Holmes A J, Kovacs K L, Murrell J C. Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch Microbiol. 1997;168:493–503. - PubMed
-
- Bowman J P, McCammon S A, Skerratt J H. Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology. 1997;143:1451–1459. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

