The ald gene, encoding a coenzyme A-acylating aldehyde dehydrogenase, distinguishes Clostridium beijerinckii and two other solvent-producing clostridia from Clostridium acetobutylicum
- PMID: 10543811
- PMCID: PMC91669
- DOI: 10.1128/AEM.65.11.4973-4980.1999
The ald gene, encoding a coenzyme A-acylating aldehyde dehydrogenase, distinguishes Clostridium beijerinckii and two other solvent-producing clostridia from Clostridium acetobutylicum
Abstract
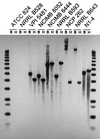
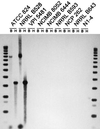
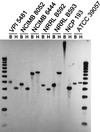
The coenzyme A (CoA)-acylating aldehyde dehydrogenase (ALDH) catalyzes a key reaction in the acetone- and butanol (solvent)-producing clostridia. It reduces acetyl-CoA and butyryl-CoA to the corresponding aldehydes, which are then reduced by alcohol dehydrogenase (ADH) to form ethanol and 1-butanol. The ALDH of Clostridium beijerinckii NRRL B593 was purified. It had no ADH activity, was NAD(H) specific, and was more active with butyraldehyde than with acetaldehyde. The N-terminal amino acid sequence of the purified ALDH was determined. The open reading frame preceding the ctfA gene (encoding a subunit of the solvent-forming CoA transferase) of C. beijerinckii NRRL B593 was identified as the structural gene (ald) for the ALDH. The ald gene encodes a polypeptide of 468 amino acid residues with a calculated M(r) of 51, 353. The position of the ald gene in C. beijerinckii NRRL B593 corresponded to that of the aad/adhE gene (encoding an aldehyde-alcohol dehydrogenase) of Clostridium acetobutylicum ATCC 824 and DSM 792. In Southern analyses, a probe derived from the C. acetobutylicum aad/adhE gene did not hybridize to restriction fragments of the genomic DNAs of C. beijerinckii and two other species of solvent-producing clostridia. In contrast, a probe derived from the C. beijerinckii ald gene hybridized to restriction fragments of the genomic DNA of three solvent-producing species but not to those of C. acetobutylicum, indicating a key difference among the solvent-producing clostridia. The amino acid sequence of the ALDH of C. beijerinckii NRRL B593 was most similar (41% identity) to those of the eutE gene products (CoA-acylating ALDHs) of Salmonella typhimurium and Escherichia coli, whereas it was about 26% identical to the ALDH domain of the aldehyde-alcohol dehydrogenases of C. acetobutylicum, E. coli, Lactococcus lactis, and amitochondriate protozoa. The predicted secondary structure of the C. beijerinckii ALDH suggests the presence of an atypical Rossmann fold for NAD(+) binding. A comparison of the proposed catalytic pockets of the CoA-dependent and CoA-independent ALDHs identified 6 amino acids that may contribute to interaction with CoA.
Figures


References
-
- Bertram J, Kuhn A, Dürre P. Tn916-induced mutants of Clostridium acetobutylicumdefective in regulation of solvent formation. Arch Microbiol. 1990;153:373–377.
-
- Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Burton R M, Stadtman E R. The oxidation of acetaldehyde to acetyl coenzyme A. J Biol Chem. 1953;20:873–890. - PubMed
-
- Chen J-S. Properties of acid- and solvent-forming enzymes of clostridia. In: Woods D R, editor. The clostridia and biotechnology. Stoneham, Mass: Butterworth-Heinemann; 1993. pp. 51–76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

