Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice
- PMID: 10545522
- PMCID: PMC409819
- DOI: 10.1172/JCI7377
Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice
Erratum in
- J Clin Invest 1999 Dec;104(12):1771. Fischman DH [corrected to Fischman DA]
Abstract
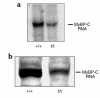
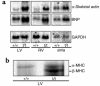
To elucidate the role of cardiac myosin-binding protein-C (MyBP-C) in myocardial structure and function, we have produced mice expressing altered forms of this sarcomere protein. The engineered mutations encode truncated forms of MyBP-C in which the cardiac myosin heavy chain-binding and titin-binding domain has been replaced with novel amino acid residues. Analogous heterozygous defects in humans cause hypertrophic cardiomyopathy. Mice that are homozygous for the mutated MyBP-C alleles express less than 10% of truncated protein in M-bands of otherwise normal sarcomeres. Homozygous mice bearing mutated MyBP-C alleles are viable but exhibit neonatal onset of a progressive dilated cardiomyopathy with prominent histopathology of myocyte hypertrophy, myofibrillar disarray, fibrosis, and dystrophic calcification. Echocardiography of homozygous mutant mice showed left ventricular dilation and reduced contractile function at birth; myocardial hypertrophy increased as the animals matured. Left-ventricular pressure-volume analyses in adult homozygous mutant mice demonstrated depressed systolic contractility with diastolic dysfunction. These data revise our understanding of the role that MyBP-C plays in myofibrillogenesis during cardiac development and indicate the importance of this protein for long-term sarcomere function and normal cardiac morphology. We also propose that mice bearing homozygous familial hypertrophic cardiomyopathy-causing mutations may provide useful tools for predicting the severity of disease that these mutations will cause in humans.
Figures





References
-
- Chien, K.R., Grace, A.A., and Hunter, J.J. 1999. Molecular and cellular biology of cardiac hypertrophy and failure. In Molecular basis of cardiovascular disease. K.R. Chien, editor. W.B. Saunders Co. Philadelphia, PA. 251–264.
-
- Rodkey, S.M., Ratliff, N.B., and Young, J.B. 1998. Cardiomyopathy and myocardial failure. In Comprehensive cardiovascular medicine. E.J. Topal, editor. Lippincott-Raven Publishers. Philadelphia, PA. 2589–2620.
-
- Watkins H, et al. Mutations in the cardiac myosin binding protein C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–437. - PubMed
-
- Bonne G, et al. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:438–440. - PubMed
-
- Niimura H, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338:1248–1257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

