Improved long-distance polymerase chain reaction for the detection of t(8;14)(q24;q32) in Burkitt's lymphomas
- PMID: 10550304
- PMCID: PMC2216451
- DOI: 10.1016/S0002-9440(10)65463-6
Improved long-distance polymerase chain reaction for the detection of t(8;14)(q24;q32) in Burkitt's lymphomas
Abstract
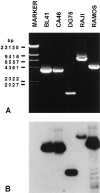
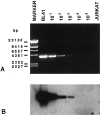
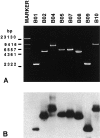
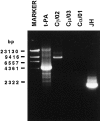
The t(8;14)(q24;q32), involving MYC gene (8q24) and the immunoglobulin heavy chain (IgH) locus (14q32), represents about 75% of all translocations in Burkitt's lymphoma (BL). Due to the great variability of the breakpoint region, a standard polymerase chain reaction assay is not sufficient for the detection of this chromosomal translocation. The availability of new and more efficient DNA polymerases that allow the amplification of genomic fragments many kilobase-pairs long, makes it possible to identify the t(8;14) in BL cells by long-distance polymerase chain reaction (LD-PCR). We have established a simplified and efficient LD-PCR for the detection of t(8;14)(q24;q32) that relies on the use of one primer specific for MYC exon II combined, in different reactions, with four primers for the IgH locus: three for the constant regions Cmu, Cgamma, and Calpha, and one for the joining region (JH). We first studied seven BL cell lines and optimized LD-PCR reaction for analysis of tumor specimens. Five of seven cell lines were positive for the t(8;14), whereas two lines derived from endemic BL were negative, as expected. Of 15 biopsies obtained from pediatric BL and subsequently analyzed, 13 (87%) were positive for the translocation detected by LD-PCR and showed a product ranging in size from 2.0 to 9.5 kb. Cmu region was involved in 6 cases, Cgamma and Calpha in 2 cases each, and JH in 3 cases. Interestingly, 2 of the tumors positive for JH showed a second, larger PCR product with the Calpha- and Cgamma-specific primer, respectively. We established that our LD-PCR method could detect 10(-3) BL cells within a population of hematopoietic cells lacking the translocation. In conclusion, our LD-PCR method represents a fast, highly sensitive, and specific tool to study sporadic BL and to detect minimal disease and residual disease in patients affected by t(8;14)-positive lymphomas.
Figures

References
-
- Shad A, Magrath I: Malignant non-Hodgkin’s lymphomas in children. Pizzo PA Poplack DG eds. Principle and Practice of Pediatric Oncology, 3d ed. 1997, :pp 545-587 Lippincott-Raven, Philadelphia
-
- Croce CM, Nowell PC: Molecular basis of B cell neoplasia. Blood 1985, 65:1-4 - PubMed
-
- Magrath I: The pathogenesis of Burkitt’s lymphoma. Adv Cancer Res 1990, 55:133-270 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

