Constitutive and inducible in vivo protein-DNA interactions at the tumor necrosis factor-alpha promoter in primary human T lymphocytes
- PMID: 10551799
- PMCID: PMC6157389
Constitutive and inducible in vivo protein-DNA interactions at the tumor necrosis factor-alpha promoter in primary human T lymphocytes
Abstract
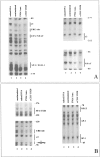
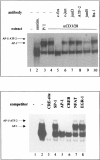
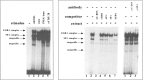
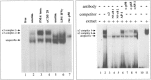
Tumor necrosis factor-alpha (TNF-alpha) is a key cytokine of lymphocytes with major regulatory functions in immunomodulation, chronic inflammation, and septic shock. However, only limited information on TNF promoter regulation in vivo in primary lymphocytes is available. To determine and compare protein-DNA interactions at the native TNF locus in primary lymphocytes, we analyzed the human TNF-alpha promoter by ligation-mediated polymerase chain reaction (LM-PCR) techniques. Accordingly, primary CD4+ T lymphocytes from peripheral blood were cultured in the presence of various stimuli and analyzed by LM-PCR. Inducible in vivo protein-DNA interactions at the TNF promoter were detected between -120 and -70 bp of the human TNF promoter relative to the transcriptional start site. This area includes binding sites for transcription factors such as ETS-1, NFAT, ATF-2/c-jun, SP-1/Egr-1, and NF-kappaB. In contrast, no protein-DNA interactions were observed at various binding sites with reported regulatory function in tumor cell lines such as the k2 element, the NFAT site at -160, the AP1 site at -50, and the SP1 site at -65. Additional mutagenesis and transfection studies demonstrated that NF-kappaB and CREB/AP-1 are important regulators of inducible TNF promoter activity in primary human T lymphocytes. These results provide novel insights into the complex regulation of TNF gene transcription in primary T lymphocytes in vivo by constitutive and inducible protein-DNA interactions that appear to be at least partially different compared to previously characterized tumor cell lines.
Figures



References
-
- Barbulescu K.; Becker C.; Schlaak J. F.; Schmitt E.; Meyer-zum-Büschenfelde K.-H.; Neurath M. F. IL-12 and IL-18 differentially activate the human IFN-gamma promoter in primary CD4+ T lymphocytes. J. Immunol. 160:3642–3649; 1998. - PubMed
-
- Barbulescu K.; Meyer-zum-Büschenfelde K.-H.; Neurath M. F. Constitutive and inducible protein/ DNA interactions of the interferon-gamma promoter in vivo in CD45RA and CD45R0 T helper subsets. Eur. J. Immunol. 27:1098–1108; 1997. - PubMed
-
- Campbell I. L.; Iscaro A.; Harrison K. IFN-gamma and tumor necrosis factor-alpha. Cytotoxicity to murine islets of Langerhans. J. Immunol. 141:2325–2329; 1988. - PubMed
-
- Di G. N.; Visvanathan K.; Lloyd A.; Wakefield D. Expression of TNF-alpha by human plasma cell in chronic inflammation. J. Leukoc. Biol. 61:667–678; 1997. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
