UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans
- PMID: 10557206
- PMCID: PMC317130
- DOI: 10.1101/gad.13.21.2774
UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans
Abstract
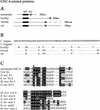
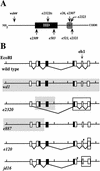
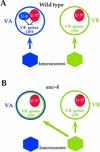
The UNC-4 homeoprotein and the Groucho-like corepressor UNC-37 specify synaptic choice in the Caenorhabditis elegans motor neuron circuit. In unc-4 mutants, VA motor neurons are miswired with inputs from interneurons normally reserved for their lineal sisters, the VB motor neurons. Here we show that UNC-4 and UNC-37 function together in VA motor neurons to repress VB-specific genes and that this activity depends on physical contact between UNC-37 and a conserved Engrailed-like repressor domain (eh1) in UNC-4. Missense mutations in the UNC-4 eh1 domain disrupt interactions between UNC-4 and UNC-37 and result in the loss of UNC-4-dependent repressor activity in vivo. A compensatory amino acid substitution in UNC-37 suppresses specific unc-4 alleles by restoring physical interactions with UNC-4 as well as UNC-4-dependent repression of VB-specific genes. We propose that repression of VB-specific genes by UNC-4 and UNC-37 is necessary for the creation of wild-type inputs to VA motor neurons. The existence of mammalian homologs of UNC-4 and UNC-37 indicates that a similar mechanism could regulate synaptic choice in the vertebrate spinal cord.
Figures




Similar articles
-
The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans.Development. 1997 May;124(9):1699-709. doi: 10.1242/dev.124.9.1699. Development. 1997. PMID: 9165118
-
UNC-4 represses CEH-12/HB9 to specify synaptic inputs to VA motor neurons in C. elegans.Genes Dev. 2007 Feb 1;21(3):332-46. doi: 10.1101/gad.1502107. Genes Dev. 2007. PMID: 17289921 Free PMC article.
-
Regulation of neurotransmitter vesicles by the homeodomain protein UNC-4 and its transcriptional corepressor UNC-37/groucho in Caenorhabditis elegans cholinergic motor neurons.J Neurosci. 2001 Mar 15;21(6):2001-14. doi: 10.1523/JNEUROSCI.21-06-02001.2001. J Neurosci. 2001. PMID: 11245684 Free PMC article.
-
Homeobox genes in Caenorhabditis elegans.Curr Opin Genet Dev. 1993 Apr;3(2):275-7. doi: 10.1016/0959-437x(93)90034-m. Curr Opin Genet Dev. 1993. PMID: 8099297 Review.
-
Mammalian Unc-13 homologues as possible regulators of neurotransmitter release.Biochem Soc Trans. 1996 Aug;24(3):661-6. doi: 10.1042/bst0240661. Biochem Soc Trans. 1996. PMID: 8878822 Review. No abstract available.
Cited by
-
Persistent engrailed expression is required to determine sensory axon trajectory, branching, and target choice.J Neurosci. 2002 Feb 1;22(3):832-41. doi: 10.1523/JNEUROSCI.22-03-00832.2002. J Neurosci. 2002. PMID: 11826113 Free PMC article.
-
brakeless is required for photoreceptor growth-cone targeting in Drosophila.Proc Natl Acad Sci U S A. 2000 May 23;97(11):5966-71. doi: 10.1073/pnas.110135297. Proc Natl Acad Sci U S A. 2000. PMID: 10811916 Free PMC article.
-
Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails.Genome Biol. 2005;6(2):R17. doi: 10.1186/gb-2005-6-2-r17. Epub 2005 Jan 31. Genome Biol. 2005. PMID: 15693946 Free PMC article.
-
Diversification of C. elegans Motor Neuron Identity via Selective Effector Gene Repression.Neuron. 2017 Jan 4;93(1):80-98. doi: 10.1016/j.neuron.2016.11.036. Neuron. 2017. PMID: 28056346 Free PMC article.
-
UNC-4 antagonizes Wnt signaling to regulate synaptic choice in the C. elegans motor circuit.Development. 2012 Jun;139(12):2234-45. doi: 10.1242/dev.075184. Development. 2012. PMID: 22619391 Free PMC article.
References
-
- Appel B, Korzh V, Glasgow E, Thor S, Edlund T, Dawid IB, Eisen JS. Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development. 1995;121:4117–4125. - PubMed
-
- Brodal P. The central nervous system. Structure and Function. 2nd ed. New York, NY: Oxford University Press; 1998.
-
- Butler MH, Wall SM, Luehrsen KR, Fox GE, Hecht RM. Molecular relationships between closely related strains and species of nematodes. J Mol Evol. 1981;18:18–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
