Double-strand-break repair recombination in Escherichia coli: physical evidence for a DNA replication mechanism in vivo
- PMID: 10557215
- PMCID: PMC317119
- DOI: 10.1101/gad.13.21.2889
Double-strand-break repair recombination in Escherichia coli: physical evidence for a DNA replication mechanism in vivo
Abstract
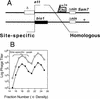
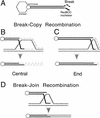
DNA double-strand-break repair (DSBR) is, in many organisms, accomplished by homologous recombination. In Escherichia coli DSBR was thought to result from breakage and reunion of parental DNA molecules, assisted by known endonucleases, the Holliday junction resolvases. Under special circumstances, for example, SOS induction, recombination forks were proposed to initiate replication. We provide physical evidence that this is a major alternative mechanism in which replication copies information from one chromosome to another generating recombinant chromosomes in normal cells in vivo. This alternative mechanism can occur independently of known Holliday junction cleaving proteins, requires DNA polymerase III, and produces recombined DNA molecules that carry newly replicated DNA. The replicational mechanism underlies about half the recombination of linear DNA in E. coli; the other half occurs by breakage and reunion, which we show requires resolvases, and is replication-independent. The data also indicate that accumulation of recombination intermediates promotes replication dramatically.
Figures





References
-
- Anderson DG, Kowalczykowski SC. The recombination hot spot chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes & Dev. 1997;11:571–581. - PubMed
-
- Bull H, Hayes S. The grpD55 locus of Escherichia coli appears to be an allele of dnaB. Mol Gen Genet. 1996;252:755–760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
