Regulation of gibberellin 20-oxidase and gibberellin 3beta-hydroxylase transcript accumulation during De-etiolation of pea seedlings
- PMID: 10557226
- PMCID: PMC59440
- DOI: 10.1104/pp.121.3.783
Regulation of gibberellin 20-oxidase and gibberellin 3beta-hydroxylase transcript accumulation during De-etiolation of pea seedlings
Abstract
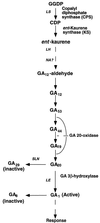
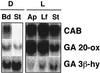
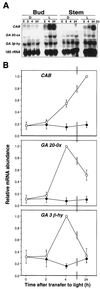
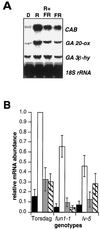
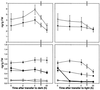
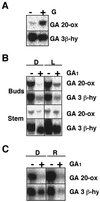
Gibberellin (GA) 20-oxidase (GA 20-ox) and GA 3beta-hydroxylase (GA 3beta-hy) are enzymes that catalyze the late steps in the formation of active GAs, and are potential control points in the regulation of GA biosynthesis by light. We have investigated the photoregulation of the GA 20-ox and GA 3beta-hy transcript levels in pea (Pisum sativum L.). The GA 20-ox transcript level was higher in light-grown seedlings than in etiolated seedlings, whereas GA 3beta-hy mRNA accumulation was higher in etiolated seedlings. However, transfer of etiolated seedlings to light led to a 5-fold increase in the expression of both transcripts 4 h after transfer. GA 20-ox mRNA accumulation is regulated by both phytochromes A and B. Transfer to light also resulted in a 6-fold decrease in GA(1) levels within 2 h. These results suggest that the light-induced drop in GA(1) level is not achieved through regulation of GA 20-ox and GA 3beta-hy mRNA accumulation. The application of exogenous GA(1) to apical buds of etiolated seedlings prior to light treatments inhibited the light-induced accumulation of both GA 20-ox and GA 3beta-hy mRNA, suggesting that negative feedback regulation is an important mechanism in the regulation of GA 20-ox and GA 3beta-hy mRNA accumulation during de-etiolation of pea seedlings.
Figures
References
-
- Ait-Ali T, Swain SM, Reid JB, Sun T-P, Kamiya Y. The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J. 1997;11:443–454. - PubMed
-
- Coruzzi G, Broghi R, Cashmore A, Chua N-H. Nucleotide sequences of two pea cDNA clones encoding the small subunit of ribulose 1,5-bisphosphate carboxylase and the major chlorophyll a/bbinding thylakoid polypeptide. J Biol Chem. 1983;258:1399–1402. - PubMed
-
- Crozier A. The Biochemistry and Physiology of Gibberellins. New York: Praeger; 1983.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

