Horizontal gene transfer and mutation: ngrol genes in the genome of Nicotiana glauca
- PMID: 10557303
- PMCID: PMC23930
- DOI: 10.1073/pnas.96.23.13229
Horizontal gene transfer and mutation: ngrol genes in the genome of Nicotiana glauca
Abstract
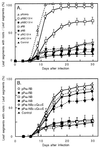
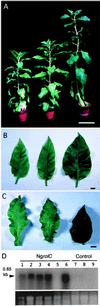
Ngrol genes (NgrolB, NgrolC, NgORF13, and NgORF14) that are similar in sequence to genes in the left transferred DNA (TL-DNA) of Agrobacterium rhizogenes have been found in the genome of untransformed plants of Nicotiana glauca. It has been suggested that a bacterial infection resulted in transformation of Ngrol genes early in the evolution of the genus Nicotiana. Although the corresponding four rol genes in TL-DNA provoked hairy-root syndrome in plants, present-day N. glauca and plants transformed with Ngrol genes did not exhibit this phenotype. Sequenced complementation analysis revealed that the NgrolB gene did not induce adventitious roots because it contained two point mutations. Single-base site-directed mutagenesis at these two positions restored the capacity for root induction to the NgrolB gene. When the NgrolB, with these two base substitutions, was positioned under the control of the cauliflower mosaic virus 35S promoter (P35S), transgenic tobacco plants exhibited morphological abnormalities that were not observed in P35S-RirolB plants. In contrast, the activity of the NgrolC gene may have been conserved after an ancient infection by bacteria. Discussed is the effect of the horizontal gene transfer of the Ngrol genes and mutations in the NgrolB gene on the phenotype of ancient plants during the evolution of N. glauca.
Figures

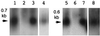

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

