Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants
- PMID: 10557305
- PMCID: PMC23932
- DOI: 10.1073/pnas.96.23.13241
Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants
Abstract
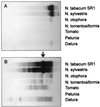
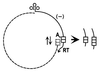
Although integration of viral DNA into host chromosomes occurs regularly in bacteria and animals, there are few reported cases in plants, and these involve insertion at only one or a few sites. Here, we report that pararetrovirus-like sequences have integrated repeatedly into tobacco chromosomes, attaining a copy number of approximately 10(3). Insertion apparently occurred by illegitimate recombination. From the sequences of 22 independent insertions recovered from a healthy plant, an 8-kilobase genome encoding a previously uncharacterized pararetrovirus that does not contain an integrase function could be assembled. Preferred boundaries of the viral inserts may correspond to recombinogenic gaps in open circular viral DNA. An unusual feature of the integrated viral sequences is a variable tandem repeat cluster, which might reflect defective genomes that preferentially recombine into plant DNA. The recurrent invasion of pararetroviral DNA into tobacco chromosomes demonstrates that viral sequences can contribute significantly to plant genome evolution.
Figures



Similar articles
-
Endogenous pararetroviral sequences in tomato (Solanum lycopersicum) and related species.BMC Plant Biol. 2007 May 21;7:24. doi: 10.1186/1471-2229-7-24. BMC Plant Biol. 2007. PMID: 17517142 Free PMC article.
-
Integration of multiple repeats of geminiviral DNA into the nuclear genome of tobacco during evolution.Proc Natl Acad Sci U S A. 1996 Jan 23;93(2):759-64. doi: 10.1073/pnas.93.2.759. Proc Natl Acad Sci U S A. 1996. PMID: 8570630 Free PMC article.
-
NTRS, a new family of highly repetitive DNAs specific for the T1 chromosome of tobacco.Chromosoma. 1997 Nov;106(6):369-79. doi: 10.1007/s004120050258. Chromosoma. 1997. PMID: 9362545
-
Viral sequences integrated into plant genomes.Annu Rev Phytopathol. 2002;40:119-36. doi: 10.1146/annurev.phyto.40.120301.105642. Epub 2002 Feb 20. Annu Rev Phytopathol. 2002. PMID: 12147756 Review.
-
Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function.Genomics Proteomics Bioinformatics. 2014 Aug;12(4):164-71. doi: 10.1016/j.gpb.2014.07.003. Epub 2014 Aug 15. Genomics Proteomics Bioinformatics. 2014. PMID: 25132181 Free PMC article. Review.
Cited by
-
Badnaviruses: The Current Global Scenario.Viruses. 2016 Jun 22;8(6):177. doi: 10.3390/v8060177. Viruses. 2016. PMID: 27338451 Free PMC article. Review.
-
Virus Latency and the Impact on Plants.Front Microbiol. 2019 Dec 6;10:2764. doi: 10.3389/fmicb.2019.02764. eCollection 2019. Front Microbiol. 2019. PMID: 31866963 Free PMC article. Review.
-
Endogenous viral sequences and their potential contribution to heritable virus resistance in plants.EMBO J. 2002 Feb 1;21(3):461-9. doi: 10.1093/emboj/21.3.461. EMBO J. 2002. PMID: 11823438 Free PMC article.
-
Endogenous pararetroviral sequences in tomato (Solanum lycopersicum) and related species.BMC Plant Biol. 2007 May 21;7:24. doi: 10.1186/1471-2229-7-24. BMC Plant Biol. 2007. PMID: 17517142 Free PMC article.
-
Rice transposable elements: a survey of 73,000 sequence-tagged-connectors.Genome Res. 2000 Jul;10(7):982-90. doi: 10.1101/gr.10.7.982. Genome Res. 2000. PMID: 10899147 Free PMC article.
References
-
- Grierson D, Covey S N. Plant Molecular Biology. New York: Chapman & Hall; 1988.
-
- Kumar A. Trends Plant Sci. 1998;3:371–374.
-
- Kiss-László Z, Hohn T. Trends Microbiol. 1996;4:480–485. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

