Feedback-inducible nuclear-receptor-driven reporter gene expression in transgenic mice
- PMID: 10557310
- PMCID: PMC23937
- DOI: 10.1073/pnas.96.23.13270
Feedback-inducible nuclear-receptor-driven reporter gene expression in transgenic mice
Abstract
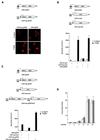
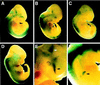
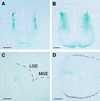
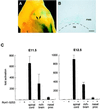
Understanding nuclear receptor signaling in vivo would be facilitated by an efficient methodology to determine where a nuclear receptor is active. Herein, we present a feedback-inducible expression system in transgenic mice to detect activated nuclear receptor effector proteins by using an inducible reporter gene. With this approach, reporter gene induction is not limited to a particular tissue, and, thus, this approach provides the opportunity for whole-animal screens. Furthermore, the effector and reporter genes are combined to generate a single strain of transgenic mice, which enables direct and rapid analysis of the offspring. The system was applied to localize sites where the retinoic acid receptor ligand-binding domain is activated in vivo. The results identify previously discovered sources of retinoids in the embryo and indicate the existence of previously undiscovered regions of retinoic acid receptor signaling in vivo. Notably, the feedback-inducible nuclear-receptor-driven assay, combined with an independent in vitro assay, provides evidence for a site of retinoid synthesis in the isthmic mesenchyme. These data illustrate the potential of feedback-inducible nuclear-receptor-driven analyses for assessing in vivo activation patterns of nuclear receptors and for analyzing pharmacological properties of natural and synthetic ligands of potential therapeutic value.
Figures
References
-
- Solomin L, Johansson C B, Zetterström R H, Bissonnette R P, Heyman R A, Olson L, Lendahl U, Frisén J, Perlmann T. Nature (London) 1998;395:398–402. - PubMed
-
- Perlmann T, Jansson L. Genes Dev. 1995;9:769–782. - PubMed
-
- Rossant J, Zirngibl R, Cado D, Shago M, Giguère V. Genes Dev. 1991;5:1333–1344. - PubMed
-
- Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. FEBS Lett. 1997;407:313–319. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

