Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage
- PMID: 10557315
- PMCID: PMC23942
- DOI: 10.1073/pnas.96.23.13300
Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage
Abstract
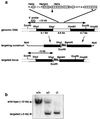
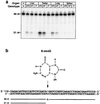
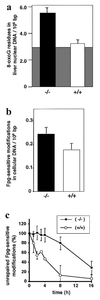
DNA damage generated by oxidant byproducts of cellular metabolism has been proposed as a key factor in cancer and aging. Oxygen free radicals cause predominantly base damage in DNA, and the most frequent mutagenic base lesion is 7,8-dihydro-8-oxoguanine (8-oxoG). This altered base can pair with A as well as C residues, leading to a greatly increased frequency of spontaneous G.C-->T.A transversion mutations in repair-deficient bacterial and yeast cells. Eukaryotic cells use a specific DNA glycosylase, the product of the OGG1 gene, to excise 8-oxoG from DNA. To assess the role of the mammalian enzyme in repair of DNA damage and prevention of carcinogenesis, we have generated homozygous ogg1(-/-) null mice. These animals are viable but accumulate abnormal levels of 8-oxoG in their genomes. Despite this increase in potentially miscoding DNA lesions, OGG1-deficient mice exhibit only a moderately, but significantly, elevated spontaneous mutation rate in nonproliferative tissues, do not develop malignancies, and show no marked pathological changes. Extracts of ogg1 null mouse tissues cannot excise the damaged base, but there is significant slow removal in vivo from proliferating cells. These findings suggest that in the absence of the DNA glycosylase, and in apparent contrast to bacterial and yeast cells, an alternative repair pathway functions to minimize the effects of an increased load of 8-oxoG in the genome and maintain a low endogenous mutation frequency.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

