Transcriptional control of the low-temperature-inducible des gene, encoding the delta5 desaturase of Bacillus subtilis
- PMID: 10559169
- PMCID: PMC94178
- DOI: 10.1128/JB.181.22.7028-7033.1999
Transcriptional control of the low-temperature-inducible des gene, encoding the delta5 desaturase of Bacillus subtilis
Abstract
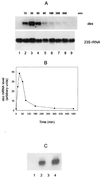
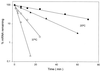
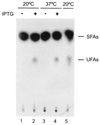
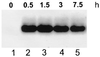
The Bacillus subtilis des gene encodes the cold-inducible Delta5 lipid desaturase involved in the formation of unsaturated fatty acids from saturated phospholipid precursors. Here, we describe the expression pattern of the des gene in response to a temperature downshift from 37 to 20 degrees C. We found that the synthesis of des mRNA is undetectable at 37 degrees C but dramatically induced upon the temperature downshift. Decay characteristics of the des transcript as well as the in vivo decay of B. subtilis bulk mRNA were investigated. The results showed that the stability of the des transcript as well as of bulk mRNA lasted substantially longer at 20 degrees C than at 37 degrees C. Functional expression of des at 37 degrees C was achieved by exchanging its promoter with the non-cold shock spac promoter. These data provide the first direct evidence that temperature-mediated control of transcription is the major mechanism regulating the mRNA levels of the B. subtilis desaturase. The present results also demonstrate that the only component of the desaturation system regulated by temperature is the desaturase enzyme.
Figures


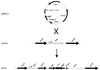

References
-
- Aguilar, P. S., and D. de Mendoza. 1998. Unpublished results.
-
- Cronan J E, Jr, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 612–636.
-
- de Mendoza D, Grau R, Cronan J E., Jr . Biosynthesis and function of membrane lipids. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 411–421.
-
- Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

