PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans
- PMID: 10559174
- PMCID: PMC94183
- DOI: 10.1128/JB.181.22.7070-7079.1999
PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans
Abstract
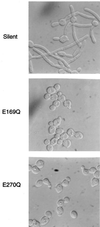
PHR1 and PHR2 encode putative glycosylphosphatidylinositol-anchored cell surface proteins of the opportunistic fungal pathogen Candida albicans. These proteins are functionally related, and their expression is modulated in relation to the pH of the ambient environment in vitro and in vivo. Deletion of either gene results in a pH-conditional defect in cell morphology and virulence. Multiple sequence alignments demonstrated a distant relationship between the Phr proteins and beta-galactosidases. Based on this alignment, site-directed mutagenesis of the putative active-site residues of Phr1p and Phr2p was conducted and two conserved glutamate residues were shown to be essential for activity. By taking advantage of the pH-conditional expression of the genes, a temporal analysis of cell wall changes was performed following a shift of the mutants from permissive to nonpermissive pH. The mutations did not grossly affect the amount of polysaccharides in the wall but did alter their distribution. The most immediate alteration to occur was a fivefold increase in the rate of cross-linking between beta-1,6-glycosylated mannoproteins and chitin. This increase was followed shortly thereafter by a decline in beta-1,3-glucan-associated beta-1, 6-glucans and, within several generations, a fivefold increase in the chitin content of the walls. The increased accumulation of chitin-linked glucans was not due to a block in subsequent processing as determined by pulse-chase analysis. Rather, the results suggest that the glucans are diverted to chitin linkage due to the inability of the mutants to establish cross-links between beta-1,6- and beta-1,3-glucans. Based on these and previously published results, it is suggested that the Phr proteins process beta-1,3-glucans and make available acceptor sites for the attachment of beta-1,6-glucans.
Figures



References
-
- Bader D E, Ring M, Huber R E. Site-directed mutagenic replacement of Glu-461 with Gln in β-galactosidase (E. coli): evidence that Glu-461 is important for activity. Biochem Biophys Res Commun. 1988;153:301–306. - PubMed
-
- Bulawa C E, Slater M, Cabib E, Au-Young J, Sburlati A, Adair W L J, Robbins P W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986;46:213–225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

