D-Allose catabolism of Escherichia coli: involvement of alsI and regulation of als regulon expression by allose and ribose
- PMID: 10559180
- PMCID: PMC94189
- DOI: 10.1128/JB.181.22.7126-7130.1999
D-Allose catabolism of Escherichia coli: involvement of alsI and regulation of als regulon expression by allose and ribose
Abstract
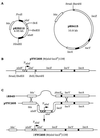
Genes involved in allose utilization of Escherichia coli K-12 are organized in at least two operons, alsRBACE and alsI, located next to each other on the chromosome but divergently transcribed. Mutants defective in alsI (allose 6-phosphate isomerase gene) and alsE (allulose 6-phosphate epimerase gene) were Als(-). Transcription of the two allose operons, measured as beta-galactosidase activity specified by alsI-lacZ(+) or alsE-lacZ(+) operon fusions, was induced by allose. Ribose also caused derepression of expression of the regulon under conditions in which ribose phosphate catabolism was impaired.
Figures
Similar articles
-
The D-allose operon of Escherichia coli K-12.J Bacteriol. 1997 Dec;179(24):7631-7. doi: 10.1128/jb.179.24.7631-7637.1997. J Bacteriol. 1997. PMID: 9401019 Free PMC article.
-
Biotransformation of Fructose to Allose by a One-Pot Reaction Using Flavonifractor plautiiD-Allulose 3-Epimerase and Clostridium thermocellum Ribose 5-Phosphate Isomerase.J Microbiol Biotechnol. 2018 Mar 28;28(3):418-424. doi: 10.4014/jmb.1709.09044. J Microbiol Biotechnol. 2018. PMID: 29316745
-
Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression.J Bacteriol. 1996 Feb;178(4):1003-11. doi: 10.1128/jb.178.4.1003-1011.1996. J Bacteriol. 1996. PMID: 8576032 Free PMC article.
-
The effects of DNA supercoiling on the expression of operons of the ilv regulon of Escherichia coli suggest a physiological rationale for divergently transcribed operons.Mol Microbiol. 2001 Mar;39(5):1109-15. doi: 10.1111/j.1365-2958.2001.02309.x. Mol Microbiol. 2001. PMID: 11251829 Review.
-
The galactose regulon of Escherichia coli.Mol Microbiol. 1993 Oct;10(2):245-51. doi: 10.1111/j.1365-2958.1993.tb01950.x. Mol Microbiol. 1993. PMID: 7934815 Review.
Cited by
-
Genome scale reconstruction of a Salmonella metabolic model: comparison of similarity and differences with a commensal Escherichia coli strain.J Biol Chem. 2009 Oct 23;284(43):29480-8. doi: 10.1074/jbc.M109.005868. Epub 2009 Aug 18. J Biol Chem. 2009. PMID: 19690172 Free PMC article.
-
Comparative metagenomic analyses reveal viral-induced shifts of host metabolism towards nucleotide biosynthesis.Microbiome. 2014 Mar 26;2(1):9. doi: 10.1186/2049-2618-2-9. Microbiome. 2014. PMID: 24666644 Free PMC article.
-
The transcriptional factors MurR and catabolite activator protein regulate N-acetylmuramic acid catabolism in Escherichia coli.J Bacteriol. 2008 Oct;190(20):6598-608. doi: 10.1128/JB.00642-08. Epub 2008 Aug 22. J Bacteriol. 2008. PMID: 18723630 Free PMC article.
-
Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042.PLoS One. 2010 Jan 20;5(1):e8801. doi: 10.1371/journal.pone.0008801. PLoS One. 2010. PMID: 20098708 Free PMC article.
-
Identification and Characterization of als Genes Involved in D-Allose Metabolism in Lineage II Strain of Listeria monocytogenes.Front Microbiol. 2018 Apr 4;9:621. doi: 10.3389/fmicb.2018.00621. eCollection 2018. Front Microbiol. 2018. PMID: 29670595 Free PMC article.
References
-
- Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
-
- Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heynecker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. - PubMed
-
- David J, Weismeyer H. Regulation of ribose metabolism in E. coli. II. Evidence for two ribose 5-phosphate isomerase activities. Biochim Biophys Acta. 1970;208:56–67. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

