Chromosomal integration of heterologous DNA in Escherichia coli with precise removal of markers and replicons used during construction
- PMID: 10559184
- PMCID: PMC94193
- DOI: 10.1128/JB.181.22.7143-7148.1999
Chromosomal integration of heterologous DNA in Escherichia coli with precise removal of markers and replicons used during construction
Abstract
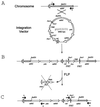
A set of vectors which facilitates the sequential integration of new functions into the Escherichia coli chromosome by homologous recombination has been developed. These vectors are based on plasmids described by Posfai et al. (J. Bacteriol. 179:4426-4428, 1997) which contain conditional replicons (pSC101 or R6K), a choice of three selectable markers (ampicillin, chloramphenicol, or kanamycin), and a single FRT site. The modified vectors contain two FRT sites which bracket a modified multiple cloning region for DNA insertion. After integration, a helper plasmid expressing the flippase (FLP) recombinase allows precise in vivo excision of the replicon and the marker used for selection. Sites are also available for temporary insertion of additional functions which can be subsequently deleted with the replicon. Only the DNA inserted into the multiple cloning sites (passenger genes and homologous fragment for targeting) and a single FRT site (68 bp) remain in the chromosome after excision. The utility of these vectors was demonstrated by integrating Zymomonas mobilis genes encoding the ethanol pathway behind the native chromosomal adhE gene in strains of E. coli K-12 and E. coli B. With these vectors, a single antibiotic selection system can be used repeatedly for the successive improvement of E. coli strains with precise deletion of extraneous genes used during construction.
Figures


References
-
- Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heynecker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. - PubMed
-
- Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

