Splicing regulatory elements within tat exon 2 of human immunodeficiency virus type 1 (HIV-1) are characteristic of group M but not group O HIV-1 strains
- PMID: 10559286
- PMCID: PMC113023
- DOI: 10.1128/JVI.73.12.9764-9772.1999
Splicing regulatory elements within tat exon 2 of human immunodeficiency virus type 1 (HIV-1) are characteristic of group M but not group O HIV-1 strains
Abstract
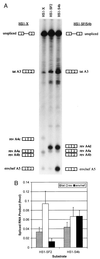
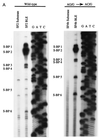
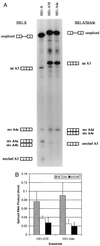
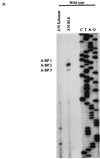
In the NL4-3 strain of human immunodeficiency virus type 1 (HIV-1), regulatory elements responsible for the relative efficiencies of alternative splicing at the tat, rev, and the env/nef 3' splice sites (A3 through A5) are contained within the region of tat exon 2 and its flanking sequences. Two elements affecting splicing of tat, rev, and env/nef mRNAs have been localized to this region. First, an exon splicing silencer (ESS2) in NL4-3, located approximately 70 nucleotides downstream from the 3' splice site used to generate tat mRNA, acts specifically to inhibit splicing at this splice site. Second, the A4b 3' splice site, which is the most downstream of the three rev 3' splice sites, also serves as an element inhibiting splicing at the env/nef 3' splice site A5. These elements are conserved in some but not all HIV-1 strains, and the effects of these sequence changes on splicing have been investigated in cell transfection and in vitro splicing assays. SF2, another clade B virus and member of the major (group M) viruses, has several sequence changes within ESS2 and uses a different rev 3' splice site. However, splicing is inhibited by the two elements similarly to NL4-3. As with the NL4-3 strain, the SF2 A4b AG dinucleotide overlaps an A5 branchpoint, and thus the inhibitory effect may result from competition of the same site for two different splicing factors. The sequence changes in ANT70C, a member of the highly divergent outlier (group O) viruses, are more extensive, and ESS2 activity in tat exon 2 is not present. Group O viruses also lack the rev 3' splice site A4b, which is conserved in all group M viruses. Mutagenesis of the most downstream rev 3' splice site of ANT70C does not increase splicing at A5, and all of the branchpoints are upstream of the two rev 3' splice sites. Thus, splicing regulatory elements in tat exon 2 which are characteristic of most group M HIV-1 strains are not present in group O HIV-1 strains.
Figures



References
-
- Aiyar A, Leis J. Modification of the megaprimer method of PCR mutagenesis: improved amplification of the final product. BioTechniques. 1993;14:366–369. - PubMed
-
- Bibollet-Ruche F, Loussert-Ajaka I, Simon F, Mboup S, Ngole E M, Saman E, Delaporte E, Peeters M. Genetic characterization of accessory genes from human immunodeficiency virus type 1 group O strains. AIDS Res Hum Retroviruses. 1998;14:951–961. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

