The residues between the two transformation effector sites of Epstein-Barr virus latent membrane protein 1 are not critical for B-lymphocyte growth transformation
- PMID: 10559303
- PMCID: PMC113040
- DOI: 10.1128/JVI.73.12.9908-9916.1999
The residues between the two transformation effector sites of Epstein-Barr virus latent membrane protein 1 are not critical for B-lymphocyte growth transformation
Abstract
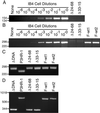
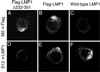
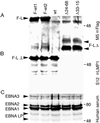
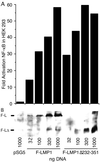
Epstein-Barr virus (EBV) latent membrane protein 1 (LMP1) is essential for EBV-mediated transformation of primary B lymphocytes. LMP1 spontaneously aggregates in the plasma membrane and enables two transformation effector sites (TES1 and TES2) within the 200-amino-acid cytoplasmic carboxyl terminus to constitutively engage the tumor necrosis factor receptor (TNFR)-associated factors TRAF1, TRAF2, TRAF3, and TRAF5 and the TNFR-associated death domain proteins TRADD and RIP, thereby activating NF-kappaB and c-Jun N-terminal kinase (JNK). To investigate the importance of the 60% of the LMP1 carboxyl terminus that lies between the TES1-TRAF and TES2-TRADD and -RIP binding sites, an EBV recombinant was made that contains a specific deletion of LMP1 codons 232 to 351. Surprisingly, the deletion mutant was similar to wild-type (wt) LMP1 EBV recombinants in its efficiency in transforming primary B lymphocytes into lymphoblastoid cell lines (LCLs). Mutant and wt EBV-transformed LCLs were similarly efficient in long-term outgrowth and in regrowth after endpoint dilution. Mutant and wt LMP1 proteins were also similar in their constitutive association with TRAF1, TRAF2, TRAF3, TRADD, and RIP. Mutant and wt EBV-transformed LCLs were similar in steady-state levels of Bcl2, JNK, and activated JNK proteins. The wt phenotype of recombinants with LMP1 codons 232 to 351 deleted further demarcates TES1 and TES2, underscores their central importance in B-lymphocyte growth transformation, and provides a new perspective on LMP1 sequence variation between TES1 and TES2.
Figures



References
-
- Baichwal V R, Sugden B. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene. 1988;2:461–467. - PubMed
-
- Brodeur S R, Cheng G, Baltimore D, Thorley L D. Localization of the major NF-κB-activating site and the sole TRAF3 binding site of LMP-1 defines two distinct signaling motifs. J Biol Chem. 1997;272:19777–19784. - PubMed
-
- Cheng G, Baltimore D. TANK, a co-inducer with TRAF2 of TNF- and CD 40L-mediated NF-κB activation. Genes Dev. 1996;10:963–973. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

