Similar interactions of the poliovirus and rhinovirus 3D polymerases with the 3' untranslated region of rhinovirus 14
- PMID: 10559308
- PMCID: PMC113045
- DOI: 10.1128/JVI.73.12.9952-9958.1999
Similar interactions of the poliovirus and rhinovirus 3D polymerases with the 3' untranslated region of rhinovirus 14
Abstract
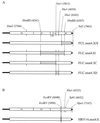
We showed previously that a human rhinovirus 14 (HRV14) 3' untranslated region (3' UTR) on a poliovirus genome was able to replicate with nearly wild-type kinetics (J. B. Rohll, D. H. Moon, D. J. Evans, and J. W. Almond, J. Virol 69:7835-7844, 1995). This enabled the HRV14 single 3' UTR stem-loop structure to be studied in combination with a sensitive reporter system, poliovirus FLC/REP, in which the capsid coding region is replaced by an in-frame chloramphemicol acetyltransferase (CAT) gene. Using such a construct, we identified a mutant (designated mut4), in which the structure and stability of the stem were predicted to be maintained, that replicated very poorly as determined by its level of CAT activity. The effect of this mutant 3' UTR on replication has been further investigated by transferring it onto the full-length cDNAs of both poliovirus type 3 (PV3) and HRV14. Virus was recovered with a parental plaque phenotype at a low frequency, indicating the acquisition of compensating changes, which sequence analysis revealed were, in both poliovirus- and rhinovirus-derived viruses, located in the active-site cleft of 3D polymerase and involved the substitution of Asn18 for Tyr. These results provide further evidence of a specific interaction between the 3' UTR of picornaviruses and the viral polymerase and also indicate similar interactions of the 3' UTR of rhinovirus with both poliovirus and rhinovirus polymerases.
Figures
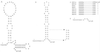




Similar articles
-
The 3' untranslated region of picornavirus RNA: features required for efficient genome replication.J Virol. 1995 Dec;69(12):7835-44. doi: 10.1128/JVI.69.12.7835-7844.1995. J Virol. 1995. PMID: 7494295 Free PMC article.
-
Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg.J Virol. 2000 Nov;74(22):10359-70. doi: 10.1128/jvi.74.22.10359-10370.2000. J Virol. 2000. PMID: 11044080 Free PMC article.
-
Analysis of the cloverleaf element in a human rhinovirus type 14/poliovirus chimera: correlation of subdomain D structure, ternary protein complex formation and virus replication.J Gen Virol. 2003 Aug;84(Pt 8):2203-2216. doi: 10.1099/vir.0.19013-0. J Gen Virol. 2003. PMID: 12867653
-
The structure-function relationship of the enterovirus 3'-UTR.Virus Res. 2009 Feb;139(2):209-16. doi: 10.1016/j.virusres.2008.07.014. Epub 2008 Aug 30. Virus Res. 2009. PMID: 18706945 Review.
-
[Structure and function of the non-coding regions of hepatitis C viral RNA].Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish.
Cited by
-
Conserved RNA secondary structures in Picornaviridae genomes.Nucleic Acids Res. 2001 Dec 15;29(24):5079-89. doi: 10.1093/nar/29.24.5079. Nucleic Acids Res. 2001. PMID: 11812840 Free PMC article.
-
Multiple functions of the nonstructural protein 3D in picornavirus infection.Front Immunol. 2024 Apr 2;15:1365521. doi: 10.3389/fimmu.2024.1365521. eCollection 2024. Front Immunol. 2024. PMID: 38629064 Free PMC article. Review.
-
An authentic 3' noncoding region is necessary for efficient poliovirus replication.J Virol. 2005 Sep;79(18):11962-73. doi: 10.1128/JVI.79.18.11962-11973.2005. J Virol. 2005. PMID: 16140772 Free PMC article.
-
Polyadenylation of genomic RNA and initiation of antigenomic RNA in a positive-strand RNA virus are controlled by the same cis-element.Nucleic Acids Res. 2006 May 31;34(10):2953-65. doi: 10.1093/nar/gkl349. Print 2006. Nucleic Acids Res. 2006. PMID: 16738134 Free PMC article.
-
Identification of a cis-acting replication element within the poliovirus coding region.J Virol. 2000 May;74(10):4590-600. doi: 10.1128/jvi.74.10.4590-4600.2000. J Virol. 2000. PMID: 10775595 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

