Multiple effects of an anti-human immunodeficiency virus nucleocapsid inhibitor on virus morphology and replication
- PMID: 10559314
- PMCID: PMC113051
- DOI: 10.1128/JVI.73.12.10000-10009.1999
Multiple effects of an anti-human immunodeficiency virus nucleocapsid inhibitor on virus morphology and replication
Abstract
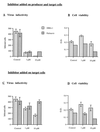
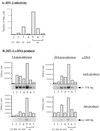
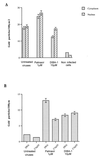
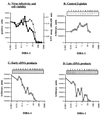
Human immunodeficiency virus type 1 nucleocapsid protein is a major structural component of the virion core and a key factor involved in proviral DNA synthesis and virus formation. 2,2'-Dithiobenzamides (DIBA-1) and related compounds that are inhibitors of NCp7 are thought to eject zinc ions from NCp7 zinc fingers, inhibiting the maturation of virion proteins. Here, we show that the presence of DIBA-1 at the time of virus formation causes morphological malformations of the virus and reduces proviral DNA synthesis. Thus, it seems that DIBA-1 is responsible for a "core-freezing effect," as shown by electron microscopy analyses. DIBA-1 can also directly interfere with the fate of the newly made proviral DNA in a manner independent of its effects on virion core formation. These data strongly suggest that nucleocapsid protein is a prime target for new compounds aimed at inhibiting human immunodeficiency virus and other retroviruses.
Figures



References
-
- Auxilien S, Keith G, Le Grice S F J, Darlix J-L. Role of post-transcriptional modifications of primer tRNALys.3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J Biol Chem. 1999;274:4412–4420. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

