Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a
- PMID: 10559320
- PMCID: PMC113057
- DOI: 10.1128/JVI.73.12.10061-10069.1999
Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a
Abstract
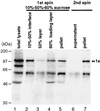
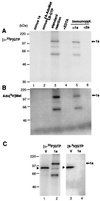
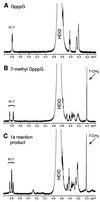
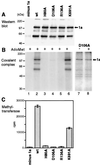
Brome mosaic virus (BMV) RNA replication is directed by two virus-encoded proteins, 1a and 2a. The amino-terminal half of 1a is a distant homolog of alphavirus nonstructural protein nsP1, which has been implicated in capping viral RNAs. In this study, we examined the enzymatic activities of BMV 1a expressed in yeast, where the protein is fully functional in RNA replication. 1a methylated GTP, dGTP, and the cap analogs GpppG and GpppA, using S-adenosylmethionine (AdoMet) as the methyl donor. Product analysis by nuclear magnetic resonance spectroscopy showed that 1a methylation was specific for guanine position 7. Additionally, 1a interacted with GTP to form a covalent 1a-m(7)GMP complex. This reaction was specific for GTP, required AdoMet, and was accompanied by transfer of (3)H-methyl from AdoMet to the covalent 1a-guanylate complex. The covalent complex could be immunoprecipitated by 1a antibodies. The 1a-m(7)GMP complex was inhibited in catalyzing further methyltransferase reactions. Mutation of conserved amino acids in the N-terminal half of 1a reduced both methyltransferase and covalent complex formation activities to very low or undetectable levels. Covalent 1a-guanylate complex formation took place in similar, AdoMet-dependent fashion in extracts of BMV-infected barley protoplasts. These results show that BMV 1a has activities similar to those of alphavirus nsP1, demonstrating conservation of these putative capping functions across a wide span of sequence divergence within the alphavirus-like superfamily. Conservation of this unusual combination of functions also supports the inference that the superfamily caps viral RNAs by an unusual pathway proceeding via a m(7)GMP intermediate.
Figures




References
-
- Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

