A hydrophobic patch in ecotropic murine leukemia virus envelope protein is the putative binding site for a critical tyrosine residue on the cellular receptor
- PMID: 10559332
- PMCID: PMC113069
- DOI: 10.1128/JVI.73.12.10164-10172.1999
A hydrophobic patch in ecotropic murine leukemia virus envelope protein is the putative binding site for a critical tyrosine residue on the cellular receptor
Abstract
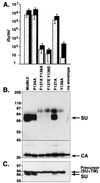
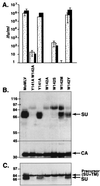
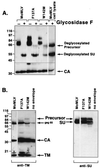
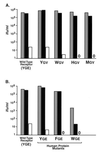
In the receptor for ecotropic murine leukemia viruses, tyrosine 235 contributes a critical hydrophobic side chain to the virus-receptor interaction (14). Here we report that tryptophan 142 in ecotropic Moloney murine leukemia virus envelope protein is essential to virus binding and infection. Replacement of tryptophan 142 by alanine or serine resulted in misfolding. However, replacement by methionine (W142M) allowed correct folding of the majority of glycoprotein molecules. W142M virus showed a marked reduction in virus binding and was almost noninfectious, suggesting that tryptophan 142 is involved in receptor binding. In contrast, W142Y virus containing a replacement of tryptophan 142 with an aromatic residue (tyrosine) was as efficient as wild-type virus in infection and binding of cells expressing the wild-type receptor. However, W142Y virus was 100-fold less efficient than wild-type virus in infection of cells expressing a mutant receptor containing tryptophan instead of the critical tyrosine. These results strongly support tryptophan 142 being an essential residue on the virus envelope protein that interacts directly with the critical hydrophobic residue at position 235 of the ecotropic receptor. Tryptophan 142 forms one side of a shallow hydrophobic pocket on the surface of the envelope protein, suggesting that it might comprise the complete putative binding site for tyrosine 235. We discuss the implications of our findings with respect to two models of the envelope protein trimer. Interestingly, both models place tryptophan 142 at the interface between adjacent subunits of the trimer.
Figures

Similar articles
-
Functional dissection of the Moloney murine leukemia virus envelope protein gp70.J Virol. 1997 Mar;71(3):2092-9. doi: 10.1128/JVI.71.3.2092-2099.1997. J Virol. 1997. PMID: 9032341 Free PMC article.
-
Second-site changes affect viability of amphotropic/ecotropic chimeric enveloped murine leukemia viruses.J Virol. 2000 Jan;74(2):899-913. doi: 10.1128/jvi.74.2.899-913.2000. J Virol. 2000. PMID: 10623753 Free PMC article.
-
The critical N-linked glycan of murine leukemia virus envelope protein promotes both folding of the C-terminal domains of the precursor polyprotein and stability of the postcleavage envelope complex.J Virol. 1997 Sep;71(9):7012-9. doi: 10.1128/JVI.71.9.7012-7019.1997. J Virol. 1997. PMID: 9261431 Free PMC article.
-
Cell targeting by murine recombinant retroviruses.Bone Marrow Transplant. 1992;9 Suppl 1:139-42. Bone Marrow Transplant. 1992. PMID: 1504656 Review.
-
Recent Advances in Bunyavirus Glycoprotein Research: Precursor Processing, Receptor Binding and Structure.Viruses. 2021 Feb 23;13(2):353. doi: 10.3390/v13020353. Viruses. 2021. PMID: 33672327 Free PMC article. Review.
Cited by
-
Maturation cleavage of the murine leukemia virus Env precursor separates the transmembrane subunits to prime it for receptor triggering.Proc Natl Acad Sci U S A. 2012 May 15;109(20):7735-40. doi: 10.1073/pnas.1118125109. Epub 2012 Apr 30. Proc Natl Acad Sci U S A. 2012. PMID: 22547812 Free PMC article.
-
Targeted entry via somatostatin receptors using a novel modified retrovirus glycoprotein that delivers genes at levels comparable to those of wild-type viral glycoproteins.J Virol. 2012 Jan;86(1):373-81. doi: 10.1128/JVI.05411-11. Epub 2011 Oct 19. J Virol. 2012. PMID: 22013043 Free PMC article.
-
Mutational Analysis of Lassa Virus Glycoprotein Highlights Regions Required for Alpha-Dystroglycan Utilization.J Virol. 2017 Aug 24;91(18):e00574-17. doi: 10.1128/JVI.00574-17. Print 2017 Sep 15. J Virol. 2017. PMID: 28679759 Free PMC article.
-
Complementation of a binding-defective retrovirus by a host cell receptor mutant.J Virol. 2004 Jun;78(11):5766-72. doi: 10.1128/JVI.78.11.5766-5772.2004. J Virol. 2004. PMID: 15140974 Free PMC article.
-
Envelope proteins containing single amino acid substitutions support a structural model of the receptor-binding domain of bovine leukemia virus surface protein.J Virol. 2002 Nov;76(21):10861-72. doi: 10.1128/jvi.76.21.10861-10872.2002. J Virol. 2002. PMID: 12368329 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

