Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity
- PMID: 10559349
- PMCID: PMC113086
- DOI: 10.1128/JVI.73.12.10310-10319.1999
Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity
Abstract
Although infection by human immunodeficiency virus (HIV) typically requires an interaction between the viral envelope glycoprotein (Env), CD4, and a chemokine receptor, CD4-independent isolates of HIV and simian immunodeficiency virus have been described. The structural basis and underlying mechanisms for this phenotype are unknown. We have derived a variant of HIV-1/IIIB, termed IIIBx, that acquired the ability to utilize CXCR4 without CD4. This virus infected CD4-negative T and B cells and fused with murine 3T3 cells that expressed human CXCR4 alone. A functional IIIBx env clone exhibited several mutations compared to the CD4-dependent HXBc2 env, including the striking loss of five glycosylation sites. By constructing env chimeras with HXBc2, the determinants for CD4 independence were shown to map outside the V1/V2 and V3 hypervariable loops, which determine chemokine receptor specificity, and at least partly within an area on the gp120 core that has been implicated in forming a conserved chemokine receptor binding site. We also identified a point mutation in the C4 domain that could render the IIIBx env clone completely CD4 dependent. Mutations in the transmembrane protein (TM) were also required for CD4 independence. Remarkably, when the V3 loop of a CCR5-tropic Env was substituted for the IIIBx Env, the resulting chimera was found to utilize CCR5 but remained CD4 independent. These findings show that Env determinants for chemokine receptor specificity are distinct from those that mediate CD4-independent use of that receptor for cell fusion and provide functional evidence for multiple steps in the interaction of Env with chemokine receptors. Combined with our observation that the conserved chemokine receptor binding site on gp120 is more exposed on the IIIBx gp120 (T. L. Hoffman, C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms, Proc. Natl. Acad. Sci. USA 96:6359-6364, 1999), the findings from this study suggest novel approaches to derive and design Envs with exposed chemokine receptor binding sites for vaccine purposes.
Figures

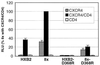
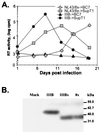

 ), as are the positions of variable loops, the gp120/gp41 cleavage site, and the TM membrane-spanning domain (msd). 8x contains a frameshift mutation at position 706 resulting in a prematurely truncated TM cytoplasmic tail. S10 contains a deletion of 50 nt, which also leads to frameshift and a prematurely truncated TM cytoplasmic tail.
), as are the positions of variable loops, the gp120/gp41 cleavage site, and the TM membrane-spanning domain (msd). 8x contains a frameshift mutation at position 706 resulting in a prematurely truncated TM cytoplasmic tail. S10 contains a deletion of 50 nt, which also leads to frameshift and a prematurely truncated TM cytoplasmic tail.




References
-
- Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. - PubMed
-
- Bandres J C, Wang Q F, O’Leary J, Baleaux F, Amara A, Hoxie J A, Zolla-Pazner S, Gorny M K. Human immunodeficiency virus (HIV) envelope binds to CXCR4 independently of CD4, and binding can be enhanced by interaction with soluble CD4 or by HIV envelope deglycosylation. J Virol. 1998;72:2500–2504. - PMC - PubMed
-
- Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11(Suppl. A):S3–S16. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

