Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection
- PMID: 10559367
- PMCID: PMC113104
- DOI: 10.1128/JVI.73.12.10489-10502.1999
Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection
Abstract
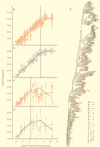
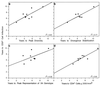
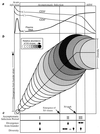
To understand the high variability of the asymptomatic interval between primary human immunodeficiency virus type 1 (HIV-1) infection and the development of AIDS, we studied the evolution of the C2-V5 region of the HIV-1 env gene and of T-cell subsets in nine men with a moderate or slow rate of disease progression. They were monitored from the time of seroconversion for a period of 6 to 12 years until the development of advanced disease in seven men. Based on the analysis of viral divergence from the founder strain, viral population diversity within sequential time points, and the outgrowth of viruses capable of utilizing the CXCR4 receptor (X4 viruses), the existence of three distinct phases within the asymptomatic interval is suggested: an early phase of variable duration during which linear increases ( approximately 1% per year) in both divergence and diversity were observed; an intermediate phase lasting an average of 1.8 years, characterized by a continued increase in divergence but with stabilization or decline in diversity; and a late phase characterized by a slowdown or stabilization of divergence and continued stability or decline in diversity. X4 variants emerged around the time of the early- to intermediate-phase transition and then achieved peak representation and began a decline around the transition between the intermediate and late phases. The late-phase transition was also associated with failure of T-cell homeostasis (defined by a downward inflection in CD3(+) T cells) and decline of CD4(+) T cells to </=200 cells/microliter. The strength of these temporal associations between viral divergence and diversity, viral coreceptor specificity, and T-cell homeostasis and subset composition supports the concept that the phases described represent a consistent pattern of viral evolution during the course of HIV-1 infection in moderate progressors. Recognition of this pattern may help explain previous conflicting data on the relationship between viral evolution and disease progression and may provide a useful framework for evaluating immune damage and recovery in untreated and treated HIV-1 infections.
Figures


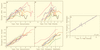

References
-
- Autran B, Carcelain G, Li T S, Blanc C, Mathez D, Tubiana R, Katlama C, Debré P, Leibowitch J. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science. 1997;277:112–116. - PubMed
-
- Berger E, Doms R, Fenyo E, Korber B, Littman D, Moore J, Sattentau Q, Schuitemaker H, Sodroski J, Weiss R. A new classification for HIV-1. Nature. 1998;391:240. - PubMed
-
- Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11(Suppl. A):S3–S16. - PubMed
-
- Berkowitz R D, Alexander S, Bare C, Linquist-Steppes V, Bogan M, Moreno M E, Gibson L, Wieder E, Kosek J, Stoddart C, McCune J M. CCR5- and CXCR4-utilizing strains of human immunodeficiency virus type 1 exhibit differential tropism and pathogenesis in vitro. J Virol. 1998;72:10108–10117. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

