Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape
- PMID: 10562274
- PMCID: PMC2156171
- DOI: 10.1083/jcb.147.4.699
Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape
Abstract
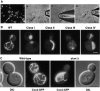
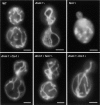
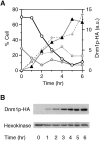
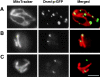
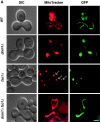
In yeast, mitochondrial division and fusion are highly regulated during growth, mating and sporulation, yet the mechanisms controlling these activities are unknown. Using a novel screen, we isolated mutants in which mitochondria lose their normal structure, and instead form a large network of interconnected tubules. These mutants, which appear defective in mitochondrial division, all carried mutations in DNM1, a dynamin-related protein that localizes to mitochondria. We also isolated mutants containing numerous mitochondrial fragments. These mutants were defective in FZO1, a gene previously shown to be required for mitochondrial fusion. Surprisingly, we found that in dnm1 fzo1 double mutants, normal mitochondrial shape is restored. Induction of Dnm1p expression in dnm1 fzo1 cells caused rapid fragmentation of mitochondria. We propose that dnm1 mutants are defective in the mitochondrial division, an activity antagonistic to fusion. Our results thus suggest that mitochondrial shape is normally controlled by a balance between division and fusion which requires Dnm1p and Fzo1p, respectively.
Figures

References
-
- Adams A., Gottschling D., Kaiser C., Stearns T. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Plainview, NY : 1997.
-
- Bereiter-Hahn J., Voth M. Dynamics of mitochondria in living cellsshape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 1994;27:198–219 . - PubMed
-
- Brachmann C.B., Davies A., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains from Saccharomyces cerevisiae 288Ca useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132 . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

