Comparative analysis of p73 and p53 regulation and effector functions
- PMID: 10562283
- PMCID: PMC2156169
- DOI: 10.1083/jcb.147.4.823
Comparative analysis of p73 and p53 regulation and effector functions
Abstract
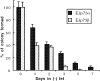
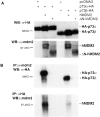
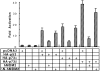
p53 is mutated in approximately 50% of human cancers, whereas mutations of the related p73 gene are rare. p73 can activate p53-responsive promoters and induce apoptosis when overexpressed in certain p53-deficient tumor cells. We show that p73 isoforms, p73alpha and p73beta, can each induce permanent growth arrest with markers of replicative senescence when overexpressed in a tetracycline-regulatable manner in human cancer cells lacking functional p53. Human homologue of mouse double minute 2 gene product (hMDM2), but not an NH(2)-terminal deletion mutant, coimmunoprecipitated with p73alpha or p73beta, and inhibited p73 transcriptional activity as with p53. In contrast to p53, ectopically expressed hemagglutinin (HA)-tagged p73 proteins were not stabilized by treatment with several DNA damaging agents. Furthermore, unlike normal p53, which increases in response to DNA damage due to enhanced protein stability in MCF7 cells, endogenous p73 protein levels were not increased in these cells under the same conditions. Thus, although p73 has an ability, comparable to that of p53, to suppress tumor cell growth in p53-deficient cells, p73 induction is regulated differently from p53. These findings suggest that the selective pressures for p53 rather than p73 inactivation in tumors may reflect their differential responses to stresses such as DNA damage, rather than their capacities to induce permanent growth arrest or apoptosis programs.
Figures
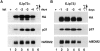



Similar articles
-
MDM2 and MDMX bind and stabilize the p53-related protein p73.Curr Biol. 1999 Jul 29-Aug 12;9(15):829-32. doi: 10.1016/s0960-9822(99)80367-4. Curr Biol. 1999. PMID: 10469568
-
Relief of p53-mediated telomerase suppression by p73.J Biol Chem. 2005 Apr 29;280(17):17329-38. doi: 10.1074/jbc.M500044200. Epub 2005 Feb 25. J Biol Chem. 2005. PMID: 15734740
-
MDM2 suppresses p73 function without promoting p73 degradation.Mol Cell Biol. 1999 May;19(5):3257-66. doi: 10.1128/MCB.19.5.3257. Mol Cell Biol. 1999. PMID: 10207051 Free PMC article.
-
Role of p73 in malignancy: tumor suppressor or oncogene?Cell Death Differ. 2002 Mar;9(3):237-45. doi: 10.1038/sj.cdd.4400995. Cell Death Differ. 2002. PMID: 11859406 Review.
-
The p53 gene family.Oncogene. 1999 Dec 13;18(53):7701-5. doi: 10.1038/sj.onc.1202955. Oncogene. 1999. PMID: 10618710 Review.
Cited by
-
p73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability.Mol Cell Biol. 2012 Jul;32(13):2336-48. doi: 10.1128/MCB.00215-12. Epub 2012 Apr 16. Mol Cell Biol. 2012. PMID: 22508983 Free PMC article.
-
p73alpha is a candidate effector in the p53 independent apoptosis pathway of cisplatin damaged primary murine colonocytes.J Clin Pathol. 2004 May;57(5):492-8. doi: 10.1136/jcp.2003.012559. J Clin Pathol. 2004. PMID: 15113856 Free PMC article.
-
Regulatory feedback loop between TP73 and TRIM32.Cell Death Dis. 2013 Jul 4;4(7):e704. doi: 10.1038/cddis.2013.224. Cell Death Dis. 2013. PMID: 23828567 Free PMC article.
-
Restoration of wild-type p53 in drug-resistant mouse breast cancer cells leads to differential gene expression, but is not sufficient to overcome the malignant phenotype.Mol Cell Biochem. 2013 Jul;379(1-2):213-27. doi: 10.1007/s11010-013-1643-5. Epub 2013 Apr 7. Mol Cell Biochem. 2013. PMID: 23564067
-
The human p73 promoter: characterization and identification of functional E2F binding sites.Neoplasia. 2002 May-Jun;4(3):195-203. doi: 10.1038/sj.neo.7900237. Neoplasia. 2002. PMID: 11988839 Free PMC article.
References
-
- Agami R., Blandino G., Oren M., Shaul Y. Interaction of c-Abl and p73α and their collaboration to induce apoptosis. Nature. 1999;399:809–813 . - PubMed
-
- Balint E., Bates S., Vousden K.H. Mdm2 binds p73α without targeting degradation. Oncogene. 1999;18:3923–3929 . - PubMed
-
- Bargonetti J., Reynisdottir I., Friedman P.N., Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 1992;6:1886–1898 . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

