Naturally processed and presented epitopes of the islet cell autoantigen IA-2 eluted from HLA-DR4
- PMID: 10562307
- PMCID: PMC409844
- DOI: 10.1172/JCI7936
Naturally processed and presented epitopes of the islet cell autoantigen IA-2 eluted from HLA-DR4
Abstract
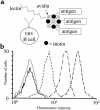
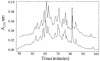
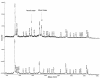
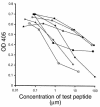
During immune responses, antigen-presenting cells (APCs) process antigens and present peptide epitopes complexed with human leukocyte antigen (HLA) molecules. CD4 cells recognize these naturally processed and presented epitopes (NPPEs) bound to HLA class II molecules. Epitope identification is important for developing diagnostic and therapeutic tools for immune-mediated diseases and providing insight into their etiology, but current approaches overlook effects of natural processing on epitope selection. We have developed a technique to identify NPPEs using mass spectrometry (MS) after antigen is targeted onto APCs using a lectin-based antigen delivery system (ADS). We applied the technique to identify NPPEs of the intracellular domain of the type 1 diabetes mellitus-associated (type 1 DM-associated) autoantigen insulinoma-associated-2 (IA-2ic), presented by HLA-DR4 (0401). IA-2ic-derived NPPEs eluted from HLA-DR4 constitute 6 sets of peptides nested around distinct core regions. Synthetic peptides based on these regions bind HLA-DR4 and elicit primary T-cell proliferation frequently in HLA-DR4-positive type 1 DM patients, but rarely in non-HLA-DR4 patients, and in none of the HLA-DR4 nondiabetic controls we tested. This flexible, direct approach identifies an HLA allele-specific map of NPPEs for any antigen, presented by any HLA class II molecule. This method should enable a greater understanding of epitope selection and lead to the generation of sensitive and specific reagents for detecting autoreactive T cells.
Figures
Comment in
-
Selecting culprits in type 1 diabetes beta-cell killing.J Clin Invest. 1999 Dec;104(11):1487-9. doi: 10.1172/JCI8826. J Clin Invest. 1999. PMID: 10587509 Free PMC article. No abstract available.
References
-
- Germain RN. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994;76:287–299. - PubMed
-
- Cramp ME, et al. Association between HLA class II genotype and spontaneous clearance of hepatitis C viraemia. J Hepatol. 1998;29:207–213. - PubMed
-
- Pardoll DM, Topalian SL. The role of CD4+ T cell responses in antitumor immunity. Curr Opin Immunol. 1998;10:588–594. - PubMed
-
- Czarnecki D, Nicholson I, Tait B, Nash C. HLA DR4 is associated with the development of multiple basal cell carcinomas and malignant melanoma. Dermatology. 1993;187:16–18. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

