Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau
- PMID: 10565899
- PMCID: PMC85833
- DOI: 10.1128/JCM.37.12.3872-3878.1999
Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau
Abstract
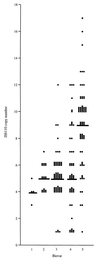
Two hundred twenty-nine consecutive isolates of Mycobacterium tuberculosis complex from patients with pulmonary tuberculosis in Guinea-Bissau, which is located in West Africa, were analyzed for clonal origin by biochemical typing and DNA fingerprinting. By using four biochemical tests (resistance to thiophene-2-carboxylic acid hydrazide, niacin production, nitrate reductase test, and pyrazinamidase test), the isolates could be assigned to five different biovars. The characteristics of four strains conformed fully with the biochemical criteria for M. bovis, while those of 85 isolates agreed with the biochemical criteria for M. tuberculosis. The remaining 140 isolates could be allocated into one of three biovars (biovars 2 to 4) representing a spectrum between the classical bovine (biovar 1) and human (biovar 5) tubercle bacilli. By using two genotyping methods, restriction fragment length polymorphism analysis with IS6110 (IS6110 RFLP analysis) and spoligotyping, the isolates could be separated into three groups (groups A to C) of the M. tuberculosis complex. Group A (n = 95), which contained the majority of classical human M. tuberculosis isolates, had large numbers of copies of IS6110 elements (mean number of copies, 9) and a distinctive spoligotyping pattern that lacked spacers 33 to 36. Isolates of the major group, group B (n = 119), had fewer IS6110 copies (mean copy number, 5) and a spoligotyping pattern that lacked spacers 7 to 9 and 39 and mainly comprised isolates of biovars 1 to 4. Group C isolates (n = 15) had one to three IS6110 copies, had a spoligotyping pattern that lacked spacers 29 to 34, and represented biovar 3 to 5 isolates. Four isolates whose biochemical characteristics conformed with those of M. bovis clustered with the group B isolates and had spoligotype patterns that differed from those previously reported for M. bovis, in that they possessed spacers 40 to 43. Interestingly, isolates of group B and, to a certain extent, also isolates of group C showed a high degree of variability in biochemical traits, despite genotypic identity in terms of IS6110 RFLP and spoligotype patterns. We hypothesize that isolates of groups B and C have their evolutionary origin in West Africa, while group A isolates are of European descent.
Figures

References
-
- Cave M D, Eisenach K D, McDermott P F, Bates J H, Crawford J T. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes. 1991;5:73–80. - PubMed
-
- David H L, Jahan M T, Jumin A, Grandry J, Lehman E H. Numerical taxonomy analysis of Mycobacterium africanum. Int J Syst Bacteriol. 1978;28:467–472.
-
- Dias F, Ghebremichael S, Hoffner S E, Martins L, Norberg R, Källenius G. Drug susceptibility in Mycobacterium tuberculosis of a sample of patients in Guinea Bissau. Tubercle. 1993;74:129–130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

