Multiplex PCR for detection and typing of porcine circoviruses
- PMID: 10565907
- PMCID: PMC85845
- DOI: 10.1128/JCM.37.12.3917-3924.1999
Multiplex PCR for detection and typing of porcine circoviruses
Erratum in
- J Clin Microbiol 2000 Apr;38(4):1707
Abstract
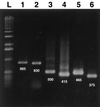
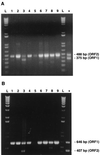
Sets of oligonucleotide primers were designed according to the sequences of the open reading frames (ORFs) ORF1 and ORF2 of the prototype nonpathogenic PK-15 strain of porcine circovirus (PCV) type 1 (PCV-1). By the PCR performed with the various primer sets, genomic DNA or RNA from other bacterial or viral pathogens of the respiratory tracts of pigs could not be amplified. A positive amplification reaction could be visualized with DNA extracted from a viral suspension containing as few as 10 viral particles per ml. No DNA fragment could be amplified from lysates of continuous porcine cell lines (PT, ST, and PFT cells) known to be negative for PCV. When tested with clinical samples from pigs, the results of the single PCR method showed nearly 93% (13 of 14 samples) correlation with histopathological and immunohistochemical findings. Interestingly, subclinical PCV infections could be detected by single PCR with clinical samples that have been submitted from animals with irrelevant cases of respiratory and/or enteric problems. On the basis of the nucleotide sequences of PCV strains (PCV-2) recently associated with outbreaks of postweaning multisystemic wasting syndrome (PWMS) in Quebec, Canada, pig farms, other primers were designed from the PCV-1 genome, and these primers failed to amplify genomic fragments specific to the ORF1 or ORF2 genes of clinical isolates associated with PWMS but amplified DNA from the PCV-1 strain. Two rapid multiplex PCR (mPCR) methods have been developed to distinguish between both genotypes of PCV. By those two mPCR methods, (i) species-specific primer pairs were used to amplify a DNA fragment of 488 bp specific for the ORF2 genes of both genotypes, whereas a 375-bp fragment was amplified from the ORF1 gene of the PCV-1 strain only, or (ii) species-specific primer pairs were used to amplify a DNA fragment of 646 bp specific for the ORF1 genes of both genotypes, whereas a 425-bp fragment was amplified from the ORF2 gene of the PCV-1 strain only. By both mPCR methods, a PCV-2 infection was demonstrated in tissues of 94.2% (33 of 35) of the sick pigs tested, in agreement with previous findings showing the close association of this new genotype of PCV with outbreaks of PMWS in Europe and North America. On the other hand, a PCV-1 infection was confirmed in only 5.7% (2 of 35) of the pigs, and confirmation of a mixed infection with PCV-2 was obtained by a single PCR with PCV-2-specific primers.
Figures


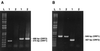

Similar articles
-
Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2.J Clin Microbiol. 2000 Jul;38(7):2494-503. doi: 10.1128/JCM.38.7.2494-2503.2000. J Clin Microbiol. 2000. PMID: 10878032 Free PMC article.
-
Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome.J Clin Microbiol. 1998 Sep;36(9):2535-41. doi: 10.1128/JCM.36.9.2535-2541.1998. J Clin Microbiol. 1998. PMID: 9705388 Free PMC article.
-
Sequence analysis of old and new strains of porcine circovirus associated with congenital tremors in pigs and their comparison with strains involved with postweaning multisystemic wasting syndrome.Can J Vet Res. 2002 Oct;66(4):217-24. Can J Vet Res. 2002. PMID: 12418776 Free PMC article.
-
Postweaning multisystemic wasting syndrome (PMWS) in pigs with particular emphasis on the causative agent, the mode of transmission, the diagnostic tools and the control measures. A review.Vet Q. 2005 Sep;27(3):105-16. doi: 10.1080/01652176.2005.9695191. Vet Q. 2005. PMID: 16238110 Review.
-
Detection of Porcine Circovirus (PCV) Using CRISPR-Cas12a/13a Coupled with Isothermal Amplification.Viruses. 2024 Sep 30;16(10):1548. doi: 10.3390/v16101548. Viruses. 2024. PMID: 39459882 Free PMC article. Review.
Cited by
-
Multiplex nested PCR compared with in situ hybridization for the differentiation of porcine circoviruses and porcine parvovirus from pigs with postweaning multisystemic wasting syndrome.Can J Vet Res. 2003 May;67(2):133-7. Can J Vet Res. 2003. PMID: 12760479 Free PMC article.
-
Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2.J Clin Microbiol. 2000 Jul;38(7):2494-503. doi: 10.1128/JCM.38.7.2494-2503.2000. J Clin Microbiol. 2000. PMID: 10878032 Free PMC article.
-
Diagnosis and differentiation of Mycoplasma hyopneumoniae and Mycoplasma hyorhinis infections in pigs by PCR amplification of the p36 and p46 genes.J Clin Microbiol. 2000 Apr;38(4):1390-6. doi: 10.1128/JCM.38.4.1390-1396.2000. J Clin Microbiol. 2000. PMID: 10747113 Free PMC article.
-
Eucaryotic expression of the nucleocapsid protein gene of porcine circovirus type 2 and use of the protein in an indirect immunofluorescence assay for serological diagnosis of postweaning multisystemic wasting syndrome in pigs.Clin Diagn Lab Immunol. 2004 Jul;11(4):736-41. doi: 10.1128/CDLI.11.4.736-741.2004. Clin Diagn Lab Immunol. 2004. PMID: 15242949 Free PMC article.
-
The emergence of porcine circovirus 2b genotype (PCV-2b) in swine in Canada.Can Vet J. 2007 Aug;48(8):811-9. Can Vet J. 2007. PMID: 17824323 Free PMC article.
References
-
- Allan G M, McNeilly F, Cassidy J P, Reilly G A C, Adair B, Ellis W A, McNulty M S. Pathogenesis of porcine circovirus: experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet Microbiol. 1995;44:49–64. - PubMed
-
- Allan G M, McNeilly F, Kennedy S, Daft B, Clarke E G, Ellis J A, Haines D M, Meeham B M, Adair B M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10:3–10. - PubMed
-
- Boevink P, Chu P W G, Keese P. Sequence of subterranean clover stunt virus DNA: affinities with the geminiviruses. Virology. 1995;207:354–361. - PubMed
-
- Clark E G. Post-weaning multisystemic wasting syndrome. Proc Am Assoc Swine Pract. 1997;28:499–501.
-
- Daft B, Nordhausen R W, Latimer K S, Niagro F D. Interstitial pneumonia and lymphadenopathy associated with circoviral infection in a six-week-old piglet. Proc Am Assoc Vet Lab Diagn. 1996;39:32.
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

