Performance criteria of DNA fingerprinting methods for typing of Helicobacter pylori isolates: experimental results and meta-analysis
- PMID: 10565934
- PMCID: PMC85883
- DOI: 10.1128/JCM.37.12.4071-4080.1999
Performance criteria of DNA fingerprinting methods for typing of Helicobacter pylori isolates: experimental results and meta-analysis
Abstract
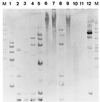
Typing systems are used to discriminate between isolates of Helicobacter pylori for epidemiological and clinical purposes. Discriminatory power and typeability are important performance criteria of typing systems. Discriminatory power refers to the ability to differentiate among unrelated isolates; it is quantitatively expressed by the discriminatory index (DI). Typeability refers to the ability of the method to provide an unambiguous result for each isolate analyzed; it is quantitatively expressed by the percentage of typeable isolates. We evaluated the discriminatory power and the typeability of the most currently used DNA fingerprinting methods for the typing of H. pylori isolates: ribotyping, PCR-based restriction fragment length polymorphism (PCR-RFLP) analysis, and random amplified polymorphism DNA (RAPD) analysis. Forty epidemiologically unrelated clinical isolates were selected to constitute a test population adapted to the evaluation of these performance criteria. A meta-analysis of typeability and discriminatory power was conducted retrospectively with raw data from published studies in which ribotyping, PCR-RFLP, RAPD, repetitive extragenic palindromic DNA sequence-based PCR (REP-PCR), or pulsed-field gel electrophoresis (PFGE) was used. Experimental results and the meta-analysis demonstrated the optimal typeability (100%) and the excellent discriminatory powers of PCR-based typing methods: RAPD analysis, DIs, 0.99 to 1; REP-PCR, DI, 0.99; and PCR-RFLP analysis, DIs, 0.70 to 0.97). Chromosome restriction-based typing methods (ribotyping and PFGE) are limited by a low typeability (12.5 to 75%) that strongly decreases their discriminatory powers: ribotyping, DI, 0.92; PFGE, DIs, 0.24 to 0.88. We do not recommend the use of ribotyping and PFGE for the typing of H. pylori isolates. We recommend the use of PCR-based methods.
Figures


References
-
- Akashi H, Hayashi T, Koizuka H, Shimoyama T, Tamura T. Strain differentiation and phylogenic relationships, in terms of base sequence of the ureB gene, of Helicobacter pylori. J Gastroenterol. 1996;31(Suppl. 9):16–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

