Biochemical analysis of the intrinsic Mcm4-Mcm6-mcm7 DNA helicase activity
- PMID: 10567526
- PMCID: PMC84885
- DOI: 10.1128/MCB.19.12.8003
Biochemical analysis of the intrinsic Mcm4-Mcm6-mcm7 DNA helicase activity
Abstract
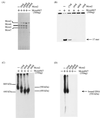
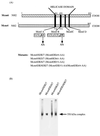
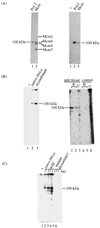
Mcm proteins play an essential role in eukaryotic DNA replication, but their biochemical functions are poorly understood. Recently, we reported that a DNA helicase activity is associated with an Mcm4-Mcm6-Mcm7 (Mcm4,6,7) complex, suggesting that this complex is involved in the initiation of DNA replication as a DNA-unwinding enzyme. In this study, we have expressed and isolated the mouse Mcm2, 4,6,7 proteins from insect cells and characterized various mutant Mcm4,6,7 complexes in which the conserved ATPase motifs of the Mcm4 and Mcm6 proteins were mutated. The activities associated with such preparations demonstrated that the DNA helicase activity is intrinsically associated with the Mcm4,6,7 complex. Biochemical analyses of these mutant Mcm4,6,7 complexes indicated that the ATP binding activity of the Mcm6 protein in the complex is critical for DNA helicase activity and that the Mcm4 protein may play a role in the single-stranded DNA binding activity of the complex. The results also indicated that the two activities of DNA helicase and single-stranded DNA binding can be separated.
Figures





References
-
- Adachi Y, Usukura J, Yanagida M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells. 1997;2:467–479. - PubMed
-
- Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Borowiec J A. DNA helicases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 545–574.
-
- Burkhart R, Schulte D, Hu D, Musahl C, Göhring F, Knippers R. Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur J Biochem. 1995;228:431–438. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
