Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion
- PMID: 10567536
- PMCID: PMC84895
- DOI: 10.1128/MCB.19.12.8103
Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion
Abstract
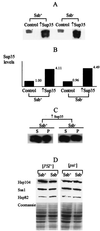
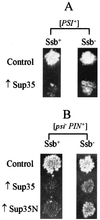
Propagation of the yeast protein-based non-Mendelian element [PSI], a prion-like form of the release factor Sup35, was shown to be regulated by the interplay between chaperone proteins Hsp104 and Hsp70. While overproduction of Hsp104 protein cures cells of [PSI], overproduction of the Ssa1 protein of the Hsp70 family protects [PSI] from the curing effect of Hsp104. Here we demonstrate that another protein of the Hsp70 family, Ssb, previously implicated in nascent polypeptide folding and protein turnover, exhibits effects on [PSI] which are opposite those of Ssa. Ssb overproduction increases, while Ssb depletion decreases, [PSI] curing by the overproduced Hsp104. Both spontaneous [PSI] formation and [PSI] induction by overproduction of the homologous or heterologous Sup35 protein are increased significantly in the strain lacking Ssb. This is the first example when inactivation of an unrelated cellular protein facilitates prion formation. Ssb is therefore playing a role in protein-based inheritance, which is analogous to the role played by the products of mutator genes in nucleic acid-based inheritance. Ssb depletion also decreases toxicity of the overproduced Sup35 and causes extreme sensitivity to the [PSI]-curing chemical agent guanidine hydrochloride. Our data demonstrate that various members of the yeast Hsp70 family have diverged from each other in regard to their roles in prion propagation and suggest that Ssb could serve as a proofreading component of the enzymatic system, which prevents formation of prion aggregates.
Figures

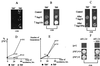

References
-
- Aigle M, Lacroute F. Genetical aspects of [URE3], a non-mitochondrial cytoplasmically inherited mutation in yeast. Mol Gen Genet. 1975;136:327–335. - PubMed
-
- Boorstein W R, Ziegelhoffer T, Craig E A. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. - PubMed
-
- Broach J R, Strathern J N, Hicks J B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979;8:121–133. - PubMed
-
- Cesareni G, Murray A H. Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In: Setlow J K, editor. Genetic engineering: principles and methods. Vol. 4. New York, N.Y: Plenum Press; 1987. pp. 13–154.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
