Coexamination of site-specific transcription factor binding and promoter activity in living cells
- PMID: 10567564
- PMCID: PMC84934
- DOI: 10.1128/MCB.19.12.8393
Coexamination of site-specific transcription factor binding and promoter activity in living cells
Abstract
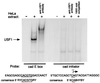
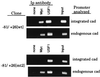
Previously, we have used a chromatin cross-linking and immunoprecipitation protocol for the analysis of Myc and USF binding to the cad promoter. The adaptation of this technique for the study of mammalian transcription factors was a big step forward in the analysis of transcription factor family member specificity, allowing for the first time a definitive knowledge of which factor binds to a promoter region under normal physiological conditions. However, due to limitations of the assay, our previous studies could not definitively prove that both Myc and USF bound to the exact same site on the cad promoter, nor could we directly correlate loss of in vivo binding of a particular factor with loss of transcriptional activity. Therefore, we have further modified the chromatin immunoprecipitation protocol to alleviate these problems. We have now shown that it is possible to coexamine growth-regulated transcriptional activity and promoter occupancy by using stably integrated promoter constructs. We show that both Myc and USF bind to the exact same E box on the cad promoter, suggesting that competition between these two factors for a single site occurs in living cells. We also find that cad promoter constructs that retain USF binding but lose Myc binding in vivo no longer display an increase in transcriptional activity in mid- to late G(1) phase of the cell cycle. Finally, we propose that cell cycle-regulated transcriptional activation of the cad promoter may be a stochastic, rather than a predetermined, process.
Figures




References
-
- Alberts A S, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. - PubMed
-
- Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA-binding by the c-Myc protein. Science. 1990;250:1149–1151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
