Distinct cellular factors regulate the c-myb promoter through its E2F element
- PMID: 10567569
- PMCID: PMC84947
- DOI: 10.1128/MCB.19.12.8442
Distinct cellular factors regulate the c-myb promoter through its E2F element
Abstract
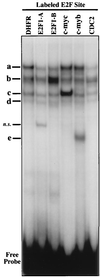
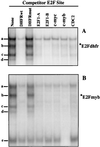
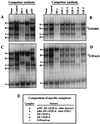
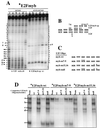
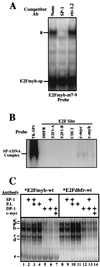
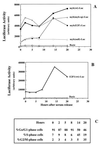
Most E2F-driven promoters are transiently activated around the G(1)/S transition. Although the promoter for the c-myb proto-oncogene harbors an E2F element, it is induced early in G(1) following entry into the cell cycle. Furthermore, this promoter remains active throughout subsequent cell cycles. Since E2F sites function as repressor elements during G(1) (due to the association of pRb with E2F factors), we investigated whether the E2F element in the c-myb promoter is regulated differently than E2F elements in promoters that are repressed during G(1). By gel shift analysis, the E2F element from the c-myb promoter was found to form a unique complex, referred to as E2Fmyb-sp, which was not observed with E2F elements from several other promoters. Antibodies to DP-1, E2F1 to -5, p107, or pRb failed to either supershift or block E2Fmyb-sp complex formation. Methylation interference experiments indicate that the DNA contact residues for the E2Fmyb-sp complex are distinct from but overlapping with residues required for the binding of E2F proteins. In addition to the identification of E2Fmyb-sp, we have found that SP-1 binds to the c-myb E2F element. Functional studies revealed that E2Fmyb-sp and/or SP-1 are required to achieve full activation of the c-myb promoter in different cell types and to maintain elevated expression of the c-myb promoter during G(1) in NIH 3T3 cells. These studies demonstrate that E2F elements can be regulated differently through the binding of unique sets of proteins.
Figures


References
-
- Adams P D, Kaelin W K. The cellular effects of E2F overexpression. Curr Top Microbiol Immunol. 1996;208:79–93. - PubMed
-
- Allen K E, de la Luna S, Kerkhoven R M, Bernards R, La Thangue N B. Distinct mechanisms of nuclear accumulation regulate the functional consequence of E2F transcription factors. J Cell Sci. 1997;110:2819–2831. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
