The Drosophila polycomb protein interacts with nucleosomal core particles In vitro via its repression domain
- PMID: 10567570
- PMCID: PMC84949
- DOI: 10.1128/MCB.19.12.8451
The Drosophila polycomb protein interacts with nucleosomal core particles In vitro via its repression domain
Abstract
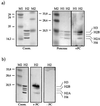
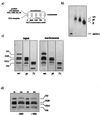
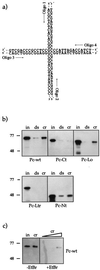
The proteins of the Polycomb group (PcG) are required for maintaining regulator genes, such as the homeotic selectors, stably and heritably repressed in appropriate developmental domains. It has been suggested that PcG proteins silence genes by creating higher-order chromatin structures at their chromosomal targets, thus preventing the interaction of components of the transcriptional machinery with their cis-regulatory elements. An unresolved issue is how higher order-structures are anchored at the chromatin base, the nucleosomal fiber. Here we show a direct biochemical interaction of a PcG protein-the Polycomb (PC) protein-with nucleosomal core particles in vitro. The main nucleosome-binding domain coincides with a region in the C-terminal part of PC previously identified as the repression domain. Our results suggest that PC, by binding to the core particle, recruits other PcG proteins to chromatin. This interaction could provide a key step in the establishment or regulation of higher-order chromatin structures.
Figures




References
-
- Alkema M J, Jacobs J, Voncken J W, Jenkins N A, Copeland N G, Satijn D P E, Otte A P, Berns A, van Lohuizen M. MPc2, a new murine homologue of the Drosophila Polycomb protein, is a member of the mouse Polycomb transcriptional repressor complex. J Mol Biol. 1997;273:993–1003. - PubMed
-
- Ausio J, Dong F, van Holde K E. Use of selectively trypsinized nucleosome core particles to analyze the role of histone “tails” in the stabilization of the nucleosome. J Mol Biol. 1989;206:451–463. - PubMed
-
- Blank T A, Becker P B. Electrostatic mechanisms of nucleosome spacing. J Mol Biol. 1995;252:305–313. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
